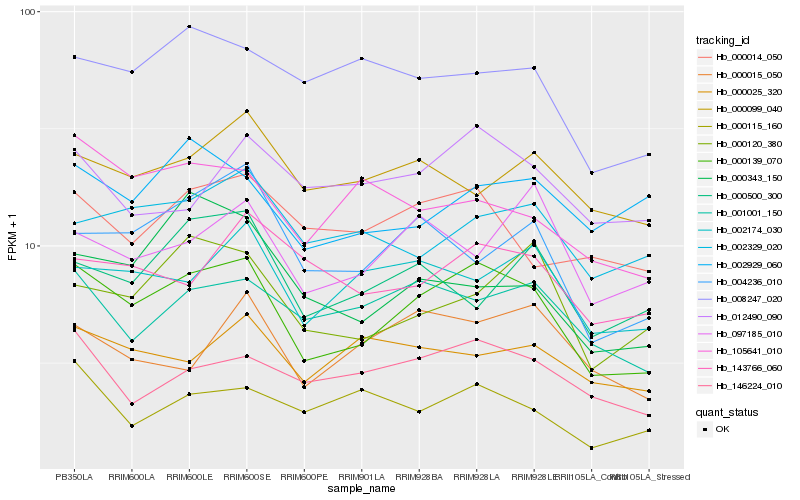
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_070 |
0.0 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 2 |
Hb_146224_010 |
0.1279007323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001001_150 |
0.1286366812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002174_030 |
0.1329587747 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 5 |
Hb_000014_050 |
0.1335602149 |
- |
- |
PREDICTED: V-type proton ATPase subunit B 2 [Elaeis guineensis] |
| 6 |
Hb_004236_010 |
0.1340329642 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 7 |
Hb_000120_380 |
0.1347822912 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 8 |
Hb_008247_020 |
0.1395033912 |
- |
- |
PREDICTED: cyclin-L1-1-like [Jatropha curcas] |
| 9 |
Hb_105641_010 |
0.1396533301 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 10 |
Hb_002329_020 |
0.1413812981 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 11 |
Hb_012490_090 |
0.1428313669 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 12 |
Hb_000099_040 |
0.1431815745 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_143766_060 |
0.1437322141 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 14 |
Hb_000025_320 |
0.1446611965 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 15 |
Hb_097185_010 |
0.1453756869 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 16 |
Hb_000015_050 |
0.1458672084 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 17 |
Hb_002929_060 |
0.1460509783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000115_160 |
0.1460510023 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 19 |
Hb_000343_150 |
0.1461963906 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 20 |
Hb_000500_300 |
0.1462225056 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |