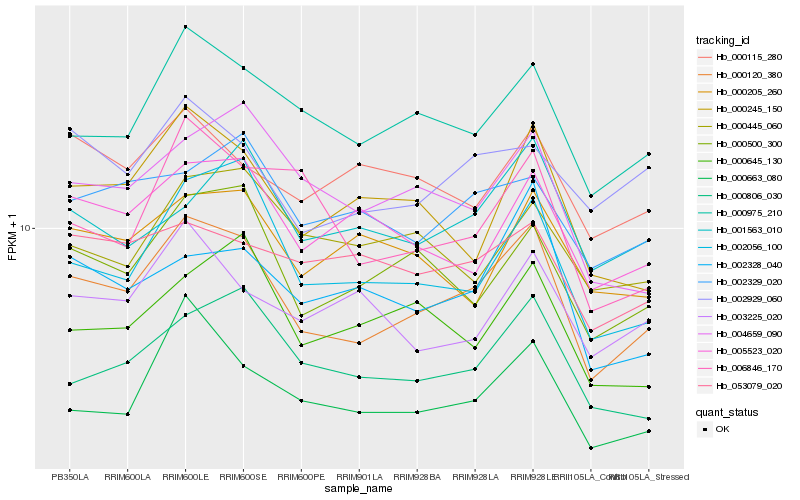
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_380 |
0.0 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 2 |
Hb_000975_210 |
0.0922177836 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 3 |
Hb_000500_300 |
0.0934388412 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |
| 4 |
Hb_002056_100 |
0.0953445036 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 5 |
Hb_000806_030 |
0.1037793256 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 6 |
Hb_000245_150 |
0.1054630839 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 7 |
Hb_000663_080 |
0.1063955245 |
transcription factor |
TF Family: HB |
Homeobox protein HAT3.1, putative [Ricinus communis] |
| 8 |
Hb_005523_020 |
0.1064377092 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 9 |
Hb_004659_090 |
0.1103843107 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 10 |
Hb_002929_060 |
0.1114500826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002328_040 |
0.113433975 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002329_020 |
0.1156433279 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 13 |
Hb_000205_260 |
0.1173866376 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 14 |
Hb_000645_130 |
0.1175994012 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 15 |
Hb_006846_170 |
0.118222233 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 16 |
Hb_003225_020 |
0.1190436725 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 17 |
Hb_000115_280 |
0.1192738612 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_001563_010 |
0.1193006319 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 19 |
Hb_053079_020 |
0.1205315093 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 20 |
Hb_000445_060 |
0.1215280924 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |