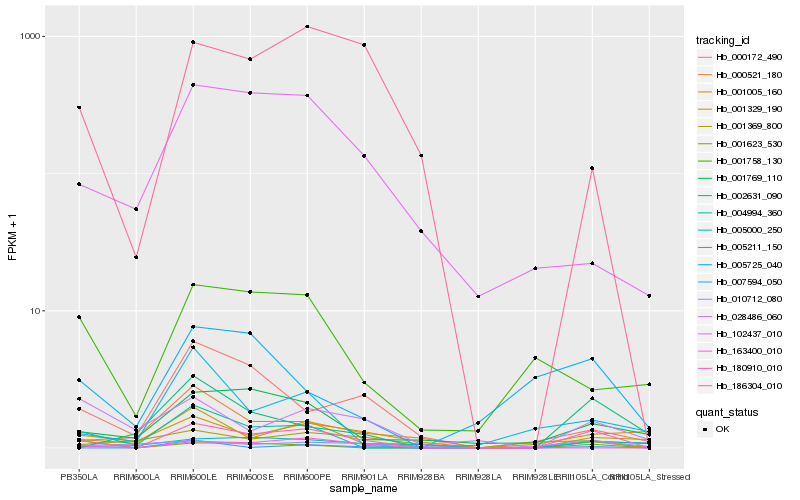
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_180910_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_186304_010 |
0.3170272916 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 3 |
Hb_001329_190 |
0.321351006 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 4 |
Hb_001769_110 |
0.323124644 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 5 |
Hb_007594_050 |
0.333165604 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_001623_530 |
0.3342681734 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 7 |
Hb_004994_360 |
0.3391416192 |
- |
- |
PREDICTED: putative ripening-related protein 1 [Populus euphratica] |
| 8 |
Hb_028486_060 |
0.3465179137 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 9 |
Hb_102437_010 |
0.3470126436 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_010712_080 |
0.3473474622 |
- |
- |
hypothetical protein AALP_AA5G019400 [Arabis alpina] |
| 11 |
Hb_002631_090 |
0.3489207867 |
- |
- |
- |
| 12 |
Hb_005000_250 |
0.3497925052 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 4, mitochondrial [Vitis vinifera] |
| 13 |
Hb_163400_010 |
0.3505475057 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 14 |
Hb_000172_490 |
0.3508177271 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 15 |
Hb_000521_180 |
0.3523645915 |
- |
- |
PREDICTED: uncharacterized protein LOC105650335 [Jatropha curcas] |
| 16 |
Hb_005725_040 |
0.35534996 |
- |
- |
- |
| 17 |
Hb_001758_130 |
0.3556719397 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 18 |
Hb_001369_800 |
0.3584315274 |
- |
- |
hypothetical protein B456_011G005000 [Gossypium raimondii] |
| 19 |
Hb_001005_160 |
0.362365681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005211_150 |
0.3636455458 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38) isoform X1 [Jatropha curcas] |