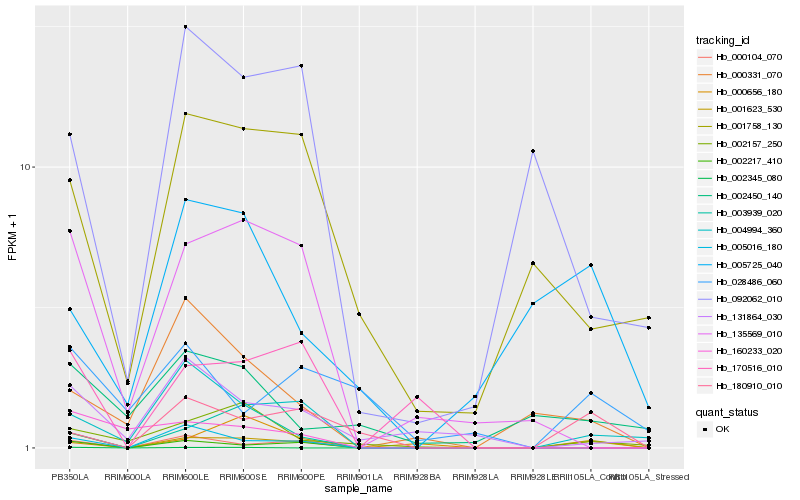
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_530 |
0.0 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 2 |
Hb_002217_410 |
0.3298620915 |
- |
- |
hypothetical protein L484_027883 [Morus notabilis] |
| 3 |
Hb_004994_360 |
0.3313177484 |
- |
- |
PREDICTED: putative ripening-related protein 1 [Populus euphratica] |
| 4 |
Hb_180910_010 |
0.3342681734 |
- |
- |
- |
| 5 |
Hb_135569_010 |
0.3458149216 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |
| 6 |
Hb_000104_070 |
0.3571059464 |
- |
- |
PREDICTED: citrate-binding protein-like [Jatropha curcas] |
| 7 |
Hb_002157_250 |
0.3571198477 |
- |
- |
Pectinesterase-4 precursor, putative [Ricinus communis] |
| 8 |
Hb_005016_180 |
0.3625842923 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 9 |
Hb_001758_130 |
0.3807722155 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 10 |
Hb_003939_020 |
0.3825634234 |
- |
- |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 11 |
Hb_131864_030 |
0.3830950721 |
transcription factor |
TF Family: C2H2 |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_005725_040 |
0.3837054096 |
- |
- |
- |
| 13 |
Hb_000656_180 |
0.3839465776 |
- |
- |
PREDICTED: uncharacterized protein LOC105631371 [Jatropha curcas] |
| 14 |
Hb_000331_070 |
0.3870842129 |
- |
- |
PREDICTED: uncharacterized protein LOC105636644 [Jatropha curcas] |
| 15 |
Hb_170516_010 |
0.3902804684 |
- |
- |
hypothetical protein MTR_001s0023 [Medicago truncatula] |
| 16 |
Hb_002345_080 |
0.3933126234 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_160233_020 |
0.3948530753 |
- |
- |
- |
| 18 |
Hb_028486_060 |
0.3963846013 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 19 |
Hb_002450_140 |
0.3982084154 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 20 |
Hb_092062_010 |
0.3993260892 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |