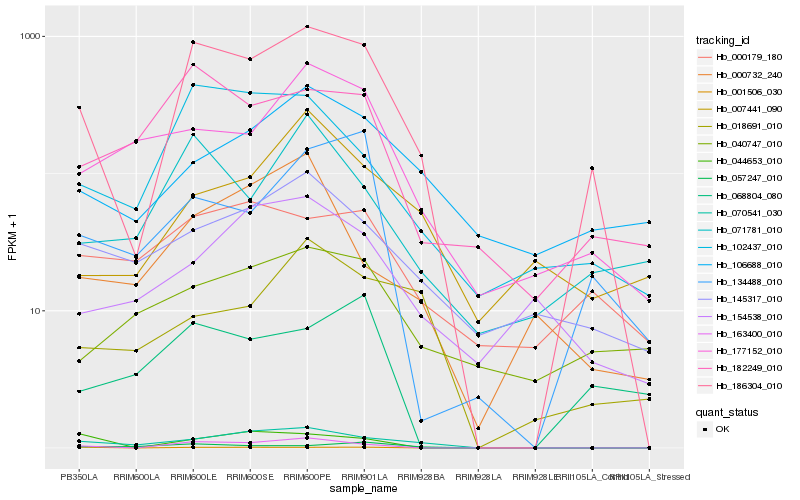
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186304_010 |
0.0 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 2 |
Hb_163400_010 |
0.2088193678 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 3 |
Hb_070541_030 |
0.2225183606 |
- |
- |
- |
| 4 |
Hb_102437_010 |
0.2336050463 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_182249_010 |
0.2440421988 |
- |
- |
- |
| 6 |
Hb_134488_010 |
0.255238111 |
- |
- |
- |
| 7 |
Hb_071781_010 |
0.2566066761 |
- |
- |
hypothetical protein POPTR_0010s13190g [Populus trichocarpa] |
| 8 |
Hb_177152_010 |
0.2570470308 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 9 |
Hb_145317_010 |
0.262634286 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 10 |
Hb_000179_180 |
0.2642663756 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 11 |
Hb_001506_030 |
0.2695418389 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_106688_010 |
0.2707599823 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 13 |
Hb_057247_010 |
0.273081575 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 14 |
Hb_018691_010 |
0.2836296164 |
- |
- |
- |
| 15 |
Hb_068804_080 |
0.2850064188 |
- |
- |
PREDICTED: protein BRICK 1 [Jatropha curcas] |
| 16 |
Hb_007441_090 |
0.2861287223 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 17 |
Hb_154538_010 |
0.2869472654 |
- |
- |
- |
| 18 |
Hb_000732_240 |
0.2886776284 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 19 |
Hb_040747_010 |
0.2898558672 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 20 |
Hb_044653_010 |
0.2921159251 |
- |
- |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |