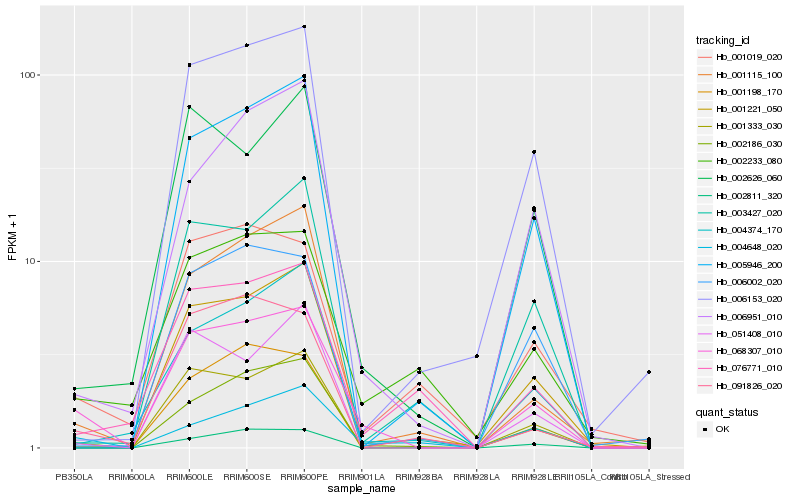
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068307_010 |
0.0 |
- |
- |
PREDICTED: phosphomannomutase [Eucalyptus grandis] |
| 2 |
Hb_002186_030 |
0.148154207 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X1 [Populus euphratica] |
| 3 |
Hb_004648_020 |
0.1871757688 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 4 |
Hb_002233_080 |
0.1922216767 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 5 |
Hb_002626_060 |
0.1939323374 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 6 |
Hb_001221_050 |
0.194275643 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 7 |
Hb_001115_100 |
0.1945785735 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 8 |
Hb_001019_020 |
0.1990785036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006951_010 |
0.1994100382 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |
| 10 |
Hb_006153_020 |
0.2001373248 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 11 |
Hb_005946_200 |
0.2029108072 |
- |
- |
hypothetical protein POPTR_0006s16210g [Populus trichocarpa] |
| 12 |
Hb_004374_170 |
0.2029517997 |
- |
- |
PREDICTED: gibberellin 20 oxidase 1-B [Jatropha curcas] |
| 13 |
Hb_091826_020 |
0.2029708406 |
- |
- |
hypothetical protein JCGZ_04975 [Jatropha curcas] |
| 14 |
Hb_001333_030 |
0.2057424287 |
- |
- |
Abscisic acid 8'-hydroxylase 1 [Theobroma cacao] |
| 15 |
Hb_051408_010 |
0.2060620303 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_003427_020 |
0.2085308716 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002811_320 |
0.2088770993 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 18 |
Hb_076771_010 |
0.2099271528 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_001198_170 |
0.2105586607 |
- |
- |
- |
| 20 |
Hb_006002_020 |
0.2125052549 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |