| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
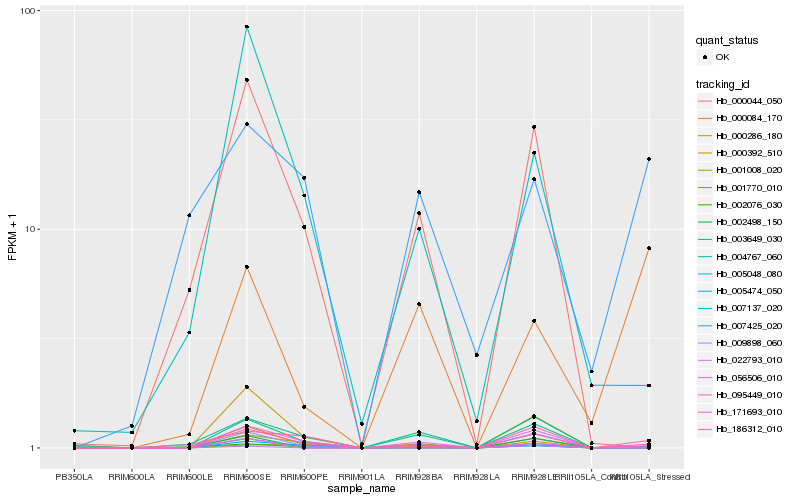
Hb_171693_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 2 |
Hb_002076_030 |
0.2285330829 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 3 |
Hb_186312_010 |
0.2682164494 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Populus euphratica] |
| 4 |
Hb_005474_050 |
0.280108498 |
- |
- |
putative leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 5 |
Hb_002498_150 |
0.2832998666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_056506_010 |
0.2867017492 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 7 |
Hb_001770_010 |
0.291985154 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 8 |
Hb_007137_020 |
0.2940025383 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 9 |
Hb_022793_010 |
0.2965648079 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 10 |
Hb_095449_010 |
0.2971590027 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein-like [Jatropha curcas] |
| 11 |
Hb_001008_020 |
0.2976740284 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_009898_060 |
0.2995478212 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 13 |
Hb_004767_060 |
0.3059995978 |
- |
- |
PREDICTED: uncharacterized protein LOC105761928 [Gossypium raimondii] |
| 14 |
Hb_000084_170 |
0.3214601471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000044_050 |
0.3222184821 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000392_510 |
0.3225742017 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 17 |
Hb_000286_180 |
0.322639567 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 18 |
Hb_003649_030 |
0.3241710867 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 19 |
Hb_007425_020 |
0.324213151 |
transcription factor |
TF Family: ERF |
PREDICTED: pathogenesis-related genes transcriptional activator PTI6-like [Jatropha curcas] |
| 20 |
Hb_005048_080 |
0.3243734964 |
- |
- |
PREDICTED: PAP-specific phosphatase HAL2-like [Jatropha curcas] |