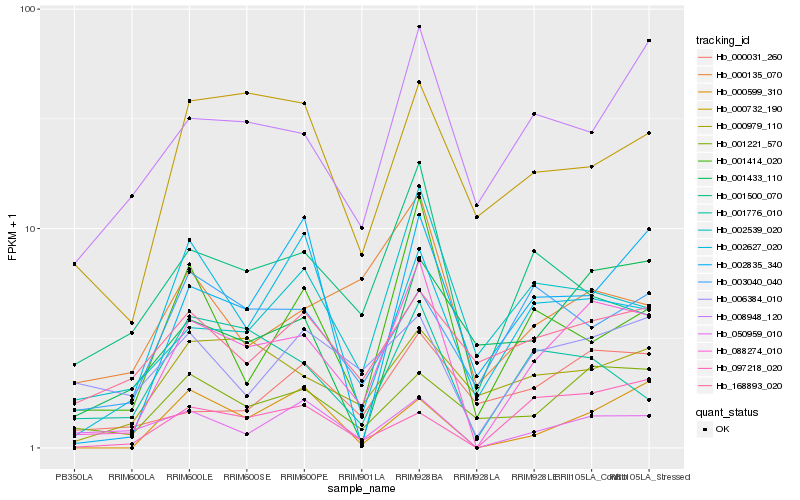
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_088274_010 |
0.0 |
- |
- |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 2 |
Hb_001221_570 |
0.216491364 |
- |
- |
- |
| 3 |
Hb_001414_020 |
0.2248273909 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_008948_120 |
0.2253547001 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_050959_010 |
0.2280196634 |
- |
- |
- |
| 6 |
Hb_002539_020 |
0.2318890548 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 7 |
Hb_001776_010 |
0.2324011225 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 8 |
Hb_000979_110 |
0.2339436131 |
- |
- |
PREDICTED: GPI mannosyltransferase 2 [Jatropha curcas] |
| 9 |
Hb_002627_020 |
0.2343778065 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 10 |
Hb_097218_020 |
0.2363894213 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 11 |
Hb_001433_110 |
0.2373187467 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006384_010 |
0.2394155353 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_000599_310 |
0.2418428693 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 14 |
Hb_001500_070 |
0.2419902165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000732_190 |
0.2425522261 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 16 |
Hb_000031_260 |
0.2430987229 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 17 |
Hb_002835_340 |
0.2435614931 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 18 |
Hb_000135_070 |
0.2437069436 |
- |
- |
PREDICTED: inositol oxygenase 1-like [Jatropha curcas] |
| 19 |
Hb_003040_040 |
0.245347168 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_168893_020 |
0.2478812303 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |