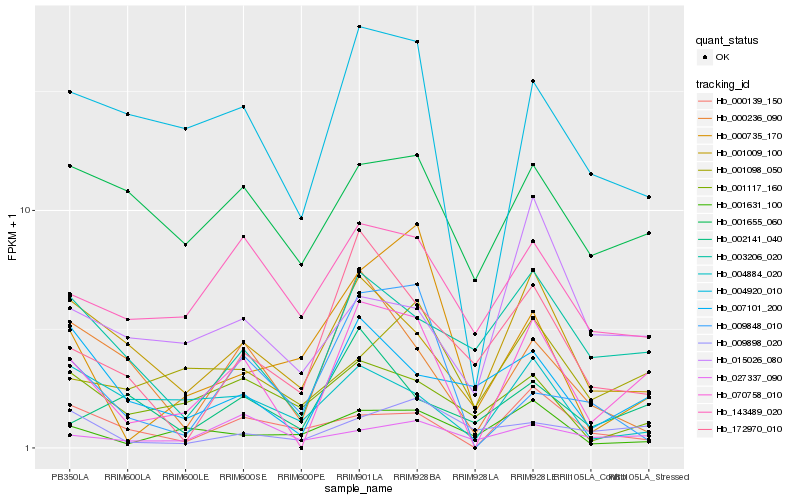
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070758_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004920_010 |
0.2376973291 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 3 |
Hb_001009_100 |
0.2658519862 |
- |
- |
- |
| 4 |
Hb_003206_020 |
0.272771031 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 5 |
Hb_001098_050 |
0.2827281854 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 6 |
Hb_000236_090 |
0.2831063475 |
- |
- |
hypothetical protein EUGRSUZ_H00030 [Eucalyptus grandis] |
| 7 |
Hb_143489_020 |
0.2849125475 |
- |
- |
- |
| 8 |
Hb_007101_200 |
0.2863670495 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 9 |
Hb_172970_010 |
0.2875970897 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 10 |
Hb_000139_150 |
0.2900566384 |
- |
- |
transposon protein, putative, Mutator sub-class [Oryza sativa Japonica Group] |
| 11 |
Hb_001631_100 |
0.2941122818 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_004884_020 |
0.2943588353 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001655_060 |
0.2971042281 |
- |
- |
PREDICTED: RNA-binding protein 28 [Jatropha curcas] |
| 14 |
Hb_001117_160 |
0.3009293019 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 15 |
Hb_000735_170 |
0.3019261962 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 16 |
Hb_027337_090 |
0.3019462423 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 17 |
Hb_002141_040 |
0.3029000295 |
- |
- |
- |
| 18 |
Hb_009848_010 |
0.3035199799 |
- |
- |
putative acyl-activating enzyme 16, chloroplastic -like protein [Gossypium arboreum] |
| 19 |
Hb_009898_020 |
0.3078222444 |
- |
- |
hypothetical protein JCGZ_23951 [Jatropha curcas] |
| 20 |
Hb_015026_080 |
0.308649417 |
- |
- |
- |