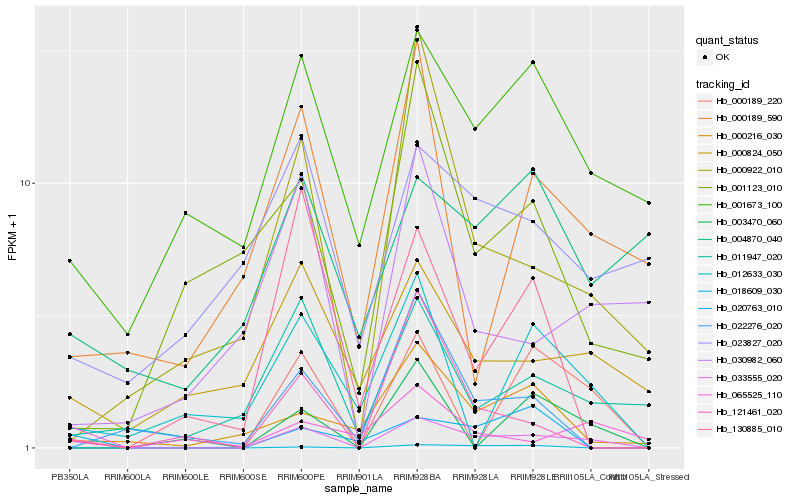
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033555_020 |
0.0 |
- |
- |
Uncharacterized protein TCM_003129 [Theobroma cacao] |
| 2 |
Hb_001673_100 |
0.3236720647 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 3 |
Hb_011947_020 |
0.3248114793 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 4 |
Hb_000922_010 |
0.325001322 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 5 |
Hb_000824_050 |
0.3327149784 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_121461_020 |
0.3368862285 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_020763_010 |
0.3424490253 |
- |
- |
polyamine oxidase, putative [Ricinus communis] |
| 8 |
Hb_000189_220 |
0.3426601468 |
- |
- |
- |
| 9 |
Hb_000216_030 |
0.3435727335 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 10 |
Hb_065525_110 |
0.346214248 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-2 [Jatropha curcas] |
| 11 |
Hb_000189_590 |
0.356076018 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 12 |
Hb_030982_060 |
0.3575249317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_023827_020 |
0.3588190151 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 14 |
Hb_130885_010 |
0.359799512 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 15 |
Hb_018609_030 |
0.3601432351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_022276_020 |
0.3624928683 |
- |
- |
- |
| 17 |
Hb_012633_030 |
0.3649321919 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 18 |
Hb_003470_060 |
0.3660159822 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 45 [Jatropha curcas] |
| 19 |
Hb_001123_010 |
0.3676152988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004870_040 |
0.3686530436 |
- |
- |
- |