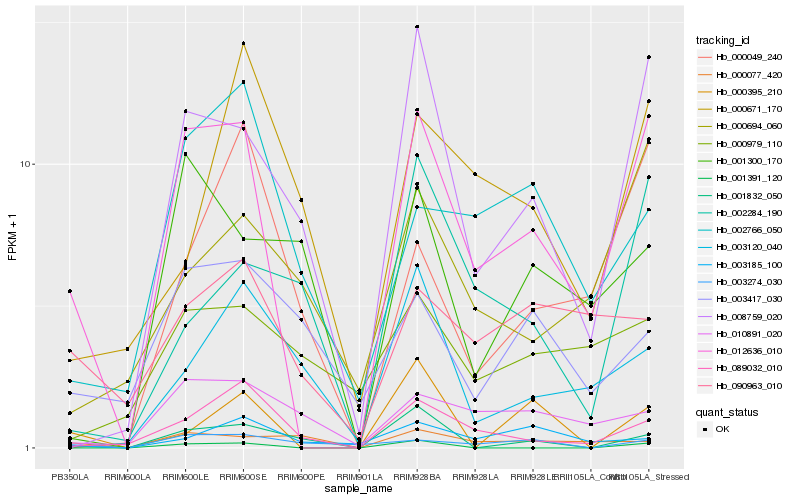
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012636_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_008759_020 |
0.2531457809 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 3 |
Hb_089032_010 |
0.2757016168 |
- |
- |
PREDICTED: flotillin-like protein 3 [Jatropha curcas] |
| 4 |
Hb_010891_020 |
0.2922383666 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 5 |
Hb_090963_010 |
0.2937612291 |
- |
- |
- |
| 6 |
Hb_002766_050 |
0.2978584195 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 7 |
Hb_000694_060 |
0.3098857214 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 8 |
Hb_000049_240 |
0.3129175385 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 9 |
Hb_003274_030 |
0.3176985838 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 10 |
Hb_001300_170 |
0.3234851775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000979_110 |
0.3246469781 |
- |
- |
PREDICTED: GPI mannosyltransferase 2 [Jatropha curcas] |
| 12 |
Hb_003185_100 |
0.3248904467 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 13 |
Hb_003417_030 |
0.3265687808 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000671_170 |
0.3280680378 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_001391_120 |
0.3287919646 |
- |
- |
Acetyl esterase, putative [Ricinus communis] |
| 16 |
Hb_000077_420 |
0.3300214241 |
- |
- |
hypothetical protein JCGZ_25815 [Jatropha curcas] |
| 17 |
Hb_000395_210 |
0.3301487577 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 18 |
Hb_003120_040 |
0.3302384006 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 19 |
Hb_002284_190 |
0.3306937576 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 20 |
Hb_001832_050 |
0.3306952977 |
- |
- |
PREDICTED: cytokinin dehydrogenase 3 [Jatropha curcas] |