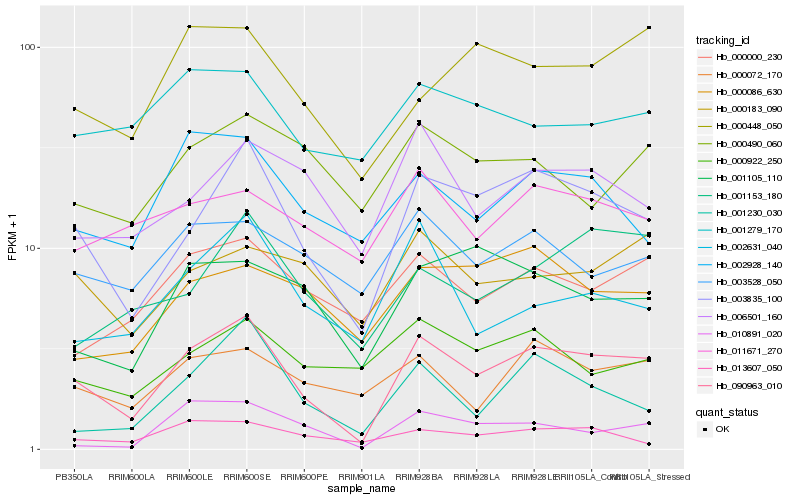
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090963_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003835_100 |
0.1114436186 |
- |
- |
PREDICTED: putative U-box domain-containing protein 50 [Jatropha curcas] |
| 3 |
Hb_000448_050 |
0.1856126907 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000072_170 |
0.1856312516 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 5 |
Hb_000000_230 |
0.187139189 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001230_030 |
0.1874444889 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X2 [Populus euphratica] |
| 7 |
Hb_002928_140 |
0.1883255632 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002631_040 |
0.189260385 |
- |
- |
hypothetical protein EUGRSUZ_J03178 [Eucalyptus grandis] |
| 9 |
Hb_001153_180 |
0.1899170913 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 10 |
Hb_006501_160 |
0.1904589132 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 11 |
Hb_003528_050 |
0.1905220672 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 12 |
Hb_000183_090 |
0.1912247401 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001105_110 |
0.1916334209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_013607_050 |
0.1969491505 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 2 [Jatropha curcas] |
| 15 |
Hb_000922_250 |
0.1972282154 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_010891_020 |
0.1976313667 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 17 |
Hb_011671_270 |
0.1990397509 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 18 |
Hb_000086_630 |
0.1991257139 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000490_060 |
0.2009833602 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 20 |
Hb_001279_170 |
0.2010187739 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |