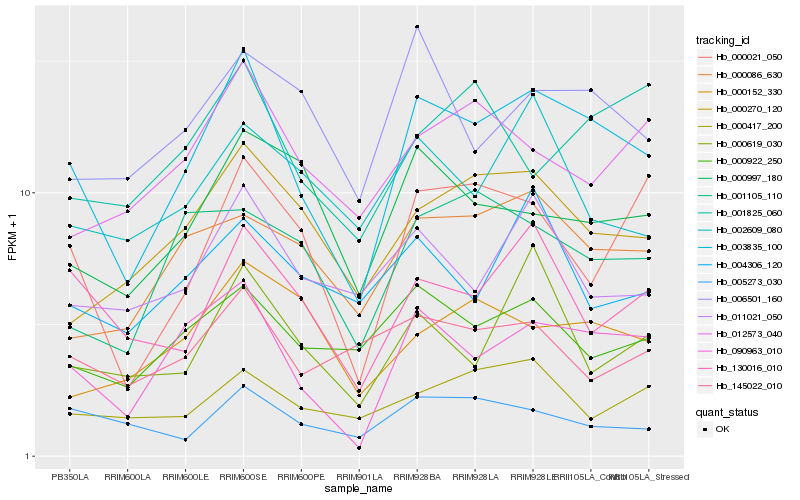
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003835_100 |
0.0 |
- |
- |
PREDICTED: putative U-box domain-containing protein 50 [Jatropha curcas] |
| 2 |
Hb_090963_010 |
0.1114436186 |
- |
- |
- |
| 3 |
Hb_130016_010 |
0.1565767082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000922_250 |
0.1594400995 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000270_120 |
0.1595411211 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 6 |
Hb_000021_050 |
0.1629707414 |
- |
- |
PREDICTED: uncharacterized protein LOC105645238 [Jatropha curcas] |
| 7 |
Hb_000086_630 |
0.1639717454 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_011021_050 |
0.1658837335 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006501_160 |
0.1678415608 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 10 |
Hb_000997_180 |
0.1689116855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001105_110 |
0.1734406252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_012573_040 |
0.1754858522 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 13 |
Hb_005273_030 |
0.1755757618 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 14 |
Hb_000619_030 |
0.1779137911 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_002609_080 |
0.1784205021 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_145022_010 |
0.1791269145 |
- |
- |
hypothetical protein JCGZ_22699 [Jatropha curcas] |
| 17 |
Hb_000417_200 |
0.1806601365 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004306_120 |
0.1809477971 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000152_330 |
0.1820854517 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 20 |
Hb_001825_060 |
0.1825259339 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |