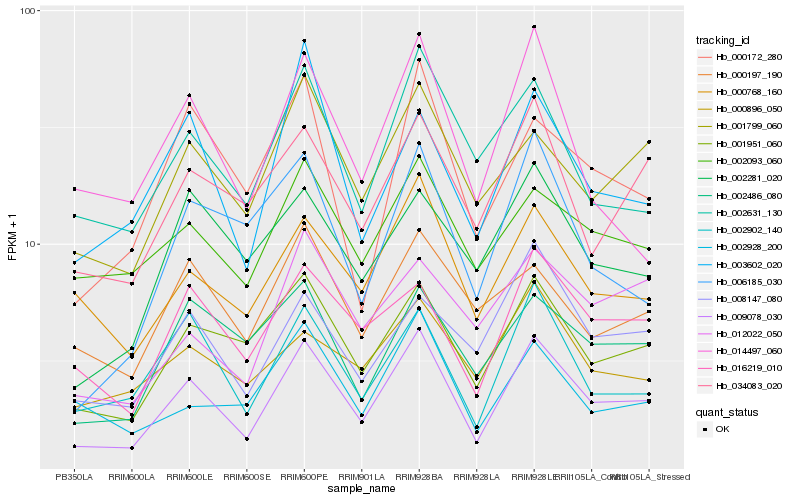
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009078_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa016992mg, partial [Prunus persica] |
| 2 |
Hb_001951_060 |
0.1114902524 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 3 |
Hb_002902_140 |
0.1186614563 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 4 |
Hb_016219_010 |
0.1254601114 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |
| 5 |
Hb_000768_160 |
0.1269110544 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 6 |
Hb_001799_060 |
0.1307483855 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 7 |
Hb_002631_130 |
0.1336829796 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 8 |
Hb_034083_020 |
0.134416598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008147_080 |
0.1369045847 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002093_060 |
0.1397585026 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_002486_080 |
0.1403763641 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 12 |
Hb_000172_280 |
0.1407778194 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 13 |
Hb_002928_200 |
0.1416184552 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 14 |
Hb_003602_020 |
0.142764393 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_014497_060 |
0.1452168343 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 16 |
Hb_002281_020 |
0.145856836 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 17 |
Hb_012022_050 |
0.1463807697 |
- |
- |
hypothetical protein EUGRSUZ_B02804 [Eucalyptus grandis] |
| 18 |
Hb_006185_030 |
0.1473046997 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 19 |
Hb_000197_190 |
0.1476668714 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000896_050 |
0.1477612295 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |