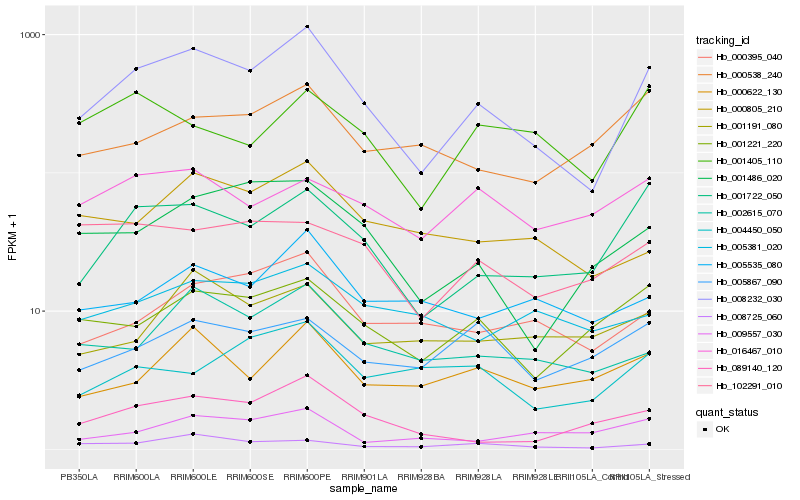
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008232_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_001722_050 |
0.1427194447 |
- |
- |
calcium binding family protein [Populus trichocarpa] |
| 3 |
Hb_002615_070 |
0.1640938139 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001486_020 |
0.1678752599 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 5 |
Hb_089140_120 |
0.1686128944 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 6 |
Hb_008725_060 |
0.1692667551 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 7 |
Hb_001221_220 |
0.1703282714 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 8 |
Hb_000395_040 |
0.1730030362 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 9 |
Hb_000805_210 |
0.1820987673 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 10 |
Hb_000622_130 |
0.1840425128 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 11 |
Hb_005535_080 |
0.1848814815 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 12 |
Hb_102291_010 |
0.1907544507 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 13 |
Hb_001191_080 |
0.1938631547 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_004450_050 |
0.1940999067 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 15 |
Hb_005867_090 |
0.1958011761 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 16 |
Hb_000538_240 |
0.1959337358 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 17 |
Hb_005381_020 |
0.1972568042 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 18 |
Hb_009557_030 |
0.1973140348 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52-like [Jatropha curcas] |
| 19 |
Hb_001405_110 |
0.1984509116 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 20 |
Hb_016467_010 |
0.1989451938 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |