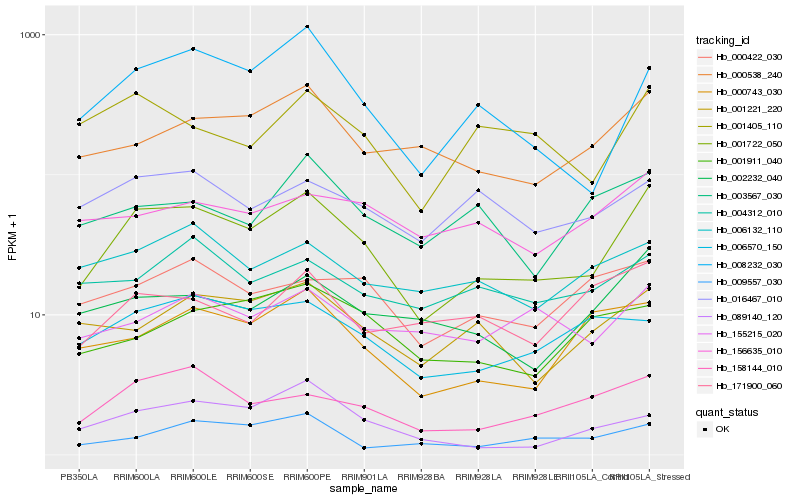
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001722_050 |
0.0 |
- |
- |
calcium binding family protein [Populus trichocarpa] |
| 2 |
Hb_008232_030 |
0.1427194447 |
- |
- |
- |
| 3 |
Hb_158144_010 |
0.152689478 |
- |
- |
- |
| 4 |
Hb_000538_240 |
0.1602994461 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 5 |
Hb_002232_040 |
0.1632160831 |
- |
- |
- |
| 6 |
Hb_001221_220 |
0.1666851095 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 7 |
Hb_000743_030 |
0.1679816737 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 8 |
Hb_001405_110 |
0.1697526651 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 9 |
Hb_006570_150 |
0.1752110021 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000422_030 |
0.1778382825 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 11 |
Hb_006132_110 |
0.1794668677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_155215_020 |
0.1801065724 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Populus euphratica] |
| 13 |
Hb_009557_030 |
0.1807034956 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52-like [Jatropha curcas] |
| 14 |
Hb_089140_120 |
0.1817968504 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 15 |
Hb_016467_010 |
0.1823494261 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 16 |
Hb_156635_010 |
0.1831199333 |
- |
- |
PREDICTED: autophagy-related protein 8f [Jatropha curcas] |
| 17 |
Hb_001911_040 |
0.1831757285 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 18 |
Hb_171900_060 |
0.1860313833 |
- |
- |
PREDICTED: uncharacterized protein LOC105643368 [Jatropha curcas] |
| 19 |
Hb_004312_010 |
0.1864241908 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003567_030 |
0.1866819324 |
- |
- |
PREDICTED: nifU-like protein 4, mitochondrial [Jatropha curcas] |