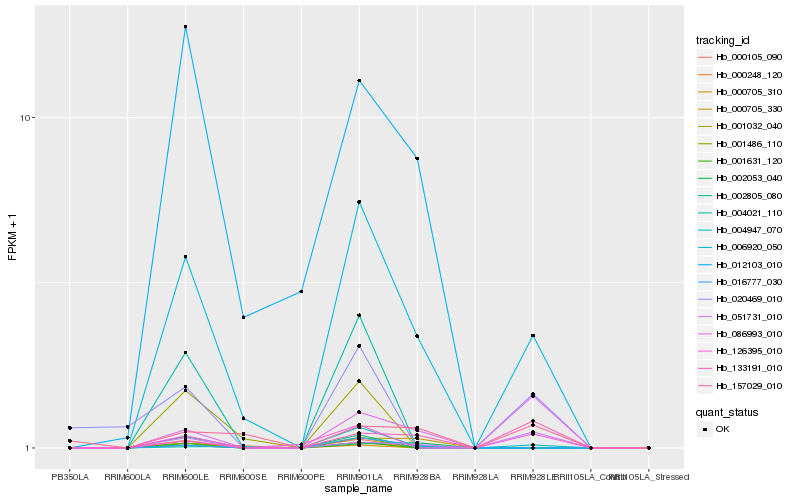
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006920_050 |
0.0 |
- |
- |
endo-1,4-beta-glucanase [Vigna radiata var. radiata] |
| 2 |
Hb_016777_030 |
0.312643762 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX11, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000248_120 |
0.3187230066 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 4 |
Hb_004947_070 |
0.3334469446 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 5 |
Hb_086993_010 |
0.3396648607 |
- |
- |
- |
| 6 |
Hb_012103_010 |
0.3397518587 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 7 |
Hb_126395_010 |
0.3539416218 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |
| 8 |
Hb_157029_010 |
0.3595511695 |
- |
- |
PREDICTED: uncharacterized protein LOC101497268 [Cicer arietinum] |
| 9 |
Hb_000705_330 |
0.36647006 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 10 |
Hb_001032_040 |
0.3698647529 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_051731_010 |
0.3771615072 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 12 |
Hb_004021_110 |
0.3806291834 |
- |
- |
- |
| 13 |
Hb_020469_010 |
0.3818717087 |
- |
- |
- |
| 14 |
Hb_133191_010 |
0.3840301293 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 15 |
Hb_002053_040 |
0.385738191 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 24 [Jatropha curcas] |
| 16 |
Hb_000705_310 |
0.3858376353 |
- |
- |
hypothetical protein POPTR_0001s03390g [Populus trichocarpa] |
| 17 |
Hb_001631_120 |
0.3859913071 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000105_090 |
0.3861842714 |
- |
- |
- |
| 19 |
Hb_002805_080 |
0.3864611284 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 20 |
Hb_001486_110 |
0.3865800624 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |