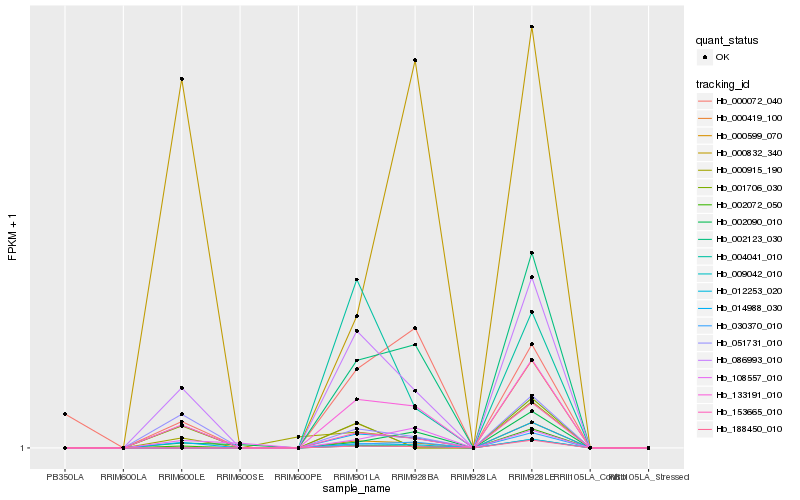
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086993_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_014988_030 |
0.0777889785 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 3 |
Hb_133191_010 |
0.0922399347 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 4 |
Hb_051731_010 |
0.1857745508 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 5 |
Hb_000915_190 |
0.272813716 |
- |
- |
- |
| 6 |
Hb_001706_030 |
0.2756514946 |
- |
- |
PREDICTED: uncharacterized protein LOC104880364 [Vitis vinifera] |
| 7 |
Hb_012253_020 |
0.2798689615 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_002072_050 |
0.28750499 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_153665_010 |
0.2963350657 |
- |
- |
PREDICTED: uncharacterized protein LOC104245945 [Nicotiana sylvestris] |
| 10 |
Hb_002090_010 |
0.2994121837 |
- |
- |
gag-pol fusion protein [Rhizophagus irregularis DAOM 197198w] |
| 11 |
Hb_002123_030 |
0.3010359747 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 12 |
Hb_000419_100 |
0.3036986639 |
- |
- |
- |
| 13 |
Hb_004041_010 |
0.3054294536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000832_340 |
0.3070727624 |
- |
- |
PREDICTED: psbP-like protein 2, chloroplastic isoform X3 [Nelumbo nucifera] |
| 15 |
Hb_000599_070 |
0.309082234 |
- |
- |
PREDICTED: patellin-4-like [Jatropha curcas] |
| 16 |
Hb_009042_010 |
0.3189427794 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_188450_010 |
0.3194098548 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_000072_040 |
0.321035512 |
- |
- |
- |
| 19 |
Hb_108557_010 |
0.3218141857 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 20 |
Hb_030370_010 |
0.3220558209 |
- |
- |
PREDICTED: uncharacterized protein LOC104896222 [Beta vulgaris subsp. vulgaris] |