| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
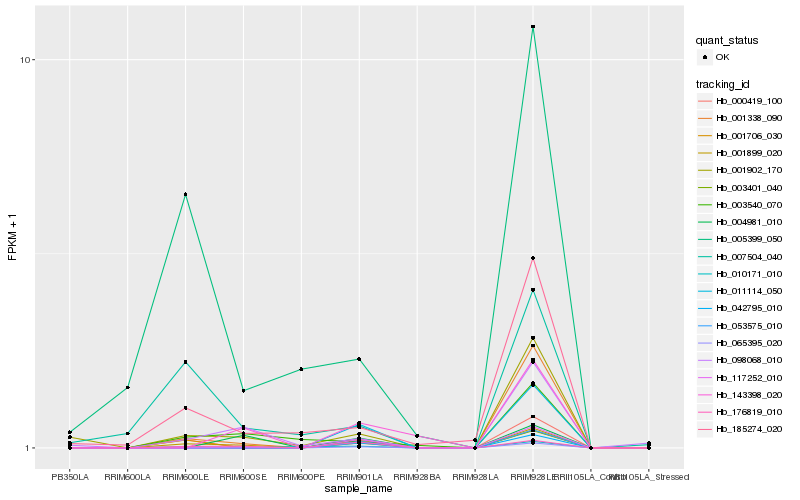
Hb_001899_020 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase TPTE2 [Jatropha curcas] |
| 2 |
Hb_176819_010 |
0.1356195808 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 3 |
Hb_000419_100 |
0.2127361734 |
- |
- |
- |
| 4 |
Hb_001706_030 |
0.2317525694 |
- |
- |
PREDICTED: uncharacterized protein LOC104880364 [Vitis vinifera] |
| 5 |
Hb_065395_020 |
0.2709567638 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 6 |
Hb_001902_170 |
0.2762710357 |
- |
- |
PREDICTED: putative glucan endo-1,3-beta-glucosidase GVI [Nicotiana sylvestris] |
| 7 |
Hb_098068_010 |
0.2887879556 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 8 [Jatropha curcas] |
| 8 |
Hb_003540_070 |
0.303129028 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 9 |
Hb_001338_090 |
0.3092987221 |
- |
- |
hypothetical protein VITISV_009275 [Vitis vinifera] |
| 10 |
Hb_185274_020 |
0.309776713 |
- |
- |
Ycf2 (chloroplast) [Andrographis paniculata] |
| 11 |
Hb_007504_040 |
0.3177250693 |
- |
- |
hypothetical chloroplast RF2 [Fragaria iinumae] |
| 12 |
Hb_004981_010 |
0.3180349857 |
- |
- |
- |
| 13 |
Hb_143398_020 |
0.3208620717 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 14 |
Hb_010171_010 |
0.3241869848 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 15 |
Hb_005399_050 |
0.3245240326 |
- |
- |
DNA-directed RNA polymerase subunit beta [Glycine soja] |
| 16 |
Hb_117252_010 |
0.3246716414 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 17 |
Hb_042795_010 |
0.324718596 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_053575_010 |
0.3250102437 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 19 |
Hb_003401_040 |
0.3252184101 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_011114_050 |
0.3254691113 |
- |
- |
- |