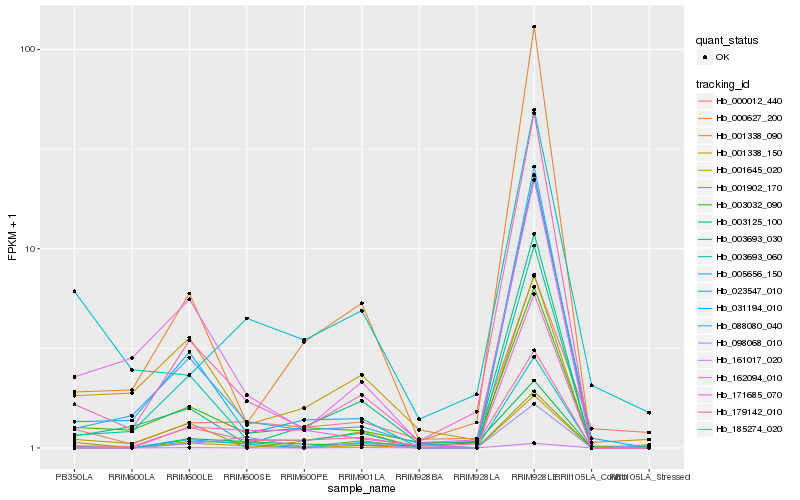
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001902_170 |
0.0 |
- |
- |
PREDICTED: putative glucan endo-1,3-beta-glucosidase GVI [Nicotiana sylvestris] |
| 2 |
Hb_161017_020 |
0.1854050013 |
- |
- |
hypothetical protein EUTSA_v10012231mg, partial [Eutrema salsugineum] |
| 3 |
Hb_098068_010 |
0.1940572767 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 8 [Jatropha curcas] |
| 4 |
Hb_185274_020 |
0.1958401505 |
- |
- |
Ycf2 (chloroplast) [Andrographis paniculata] |
| 5 |
Hb_001338_150 |
0.2059598641 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 6 |
Hb_003032_090 |
0.2101766697 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 7 |
Hb_001338_090 |
0.2127395287 |
- |
- |
hypothetical protein VITISV_009275 [Vitis vinifera] |
| 8 |
Hb_179142_010 |
0.2157245945 |
- |
- |
NADH dehydrogenase ND 2 subunit [Nicotiana tomentosiformis] |
| 9 |
Hb_005656_150 |
0.2165656344 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 10 |
Hb_031194_010 |
0.2244124221 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Arachis hypogaea] |
| 11 |
Hb_003693_030 |
0.2262969198 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 12 |
Hb_003693_060 |
0.2263046529 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 13 |
Hb_171685_070 |
0.2265343385 |
- |
- |
- |
| 14 |
Hb_003125_100 |
0.2296464534 |
- |
- |
NAD(P)-binding Rossmann-fold superfamily protein [Theobroma cacao] |
| 15 |
Hb_001645_020 |
0.2312871324 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit H [Medicago truncatula] |
| 16 |
Hb_000012_440 |
0.2316188678 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 17 |
Hb_162094_010 |
0.2318128316 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 18 |
Hb_088080_040 |
0.2329301139 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 2 A [Medicago truncatula] |
| 19 |
Hb_023547_010 |
0.2357107875 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000627_200 |
0.235869164 |
- |
- |
NADH dehydrogenase subunit I [Hevea brasiliensis] |