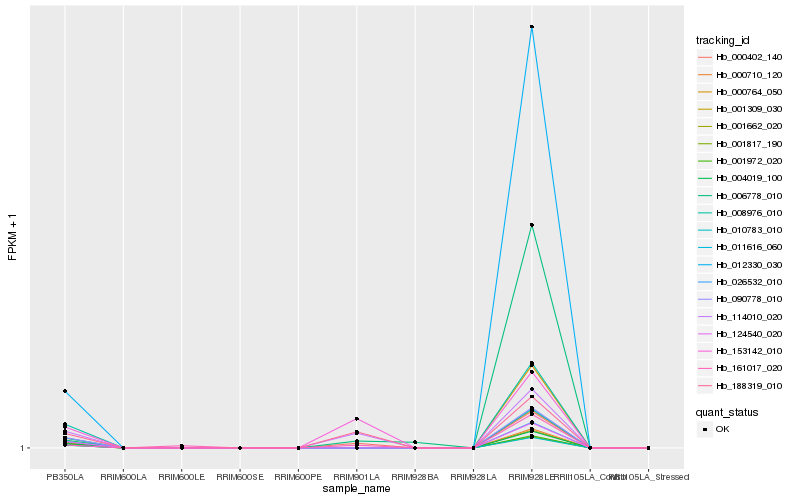
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_140 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_114010_020 |
0.1315020247 |
- |
- |
- |
| 3 |
Hb_188319_010 |
0.154538535 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 4 |
Hb_161017_020 |
0.1767371865 |
- |
- |
hypothetical protein EUTSA_v10012231mg, partial [Eutrema salsugineum] |
| 5 |
Hb_153142_010 |
0.1776703126 |
- |
- |
PREDICTED: uncharacterized protein LOC105636643 [Jatropha curcas] |
| 6 |
Hb_006778_010 |
0.2202302503 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 7 |
Hb_012330_030 |
0.2291810866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000764_050 |
0.2399789083 |
- |
- |
hypothetical protein JCGZ_09935 [Jatropha curcas] |
| 9 |
Hb_026532_010 |
0.244993245 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 10 |
Hb_001662_020 |
0.245791589 |
- |
- |
PREDICTED: F-box protein SKIP23-like isoform X3 [Eucalyptus grandis] |
| 11 |
Hb_011616_060 |
0.2458290623 |
- |
- |
PREDICTED: uncharacterized protein LOC105789623 [Gossypium raimondii] |
| 12 |
Hb_090778_010 |
0.2459156491 |
- |
- |
PREDICTED: uncharacterized protein LOC105764482 [Gossypium raimondii] |
| 13 |
Hb_124540_020 |
0.2463618349 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 14 |
Hb_010783_010 |
0.248423379 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_001817_190 |
0.2484676183 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_001972_020 |
0.2485356732 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_004019_100 |
0.2488417045 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 18 |
Hb_001309_030 |
0.2488983338 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_000710_120 |
0.249004321 |
- |
- |
hypothetical protein JCGZ_01844 [Jatropha curcas] |
| 20 |
Hb_008976_010 |
0.2491665487 |
- |
- |
- |