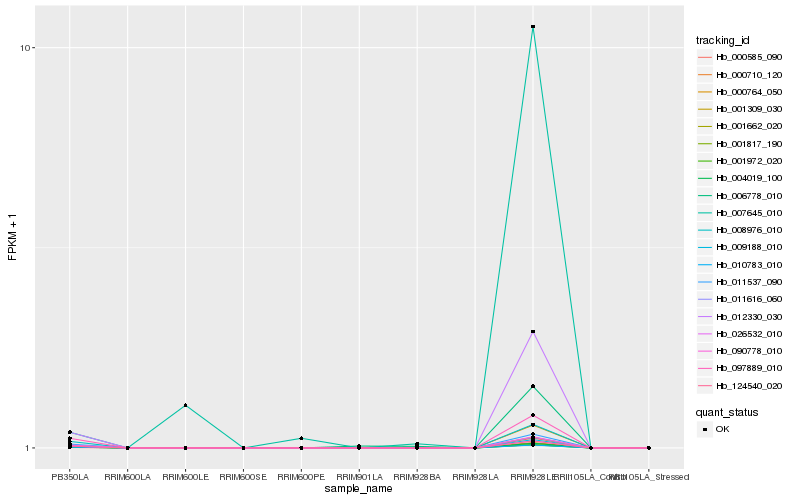
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012330_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000764_050 |
0.0450212704 |
- |
- |
hypothetical protein JCGZ_09935 [Jatropha curcas] |
| 3 |
Hb_000585_090 |
0.0962479318 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105114407 [Populus euphratica] |
| 4 |
Hb_026532_010 |
0.1328879212 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 5 |
Hb_001662_020 |
0.1349986709 |
- |
- |
PREDICTED: F-box protein SKIP23-like isoform X3 [Eucalyptus grandis] |
| 6 |
Hb_011616_060 |
0.135096794 |
- |
- |
PREDICTED: uncharacterized protein LOC105789623 [Gossypium raimondii] |
| 7 |
Hb_090778_010 |
0.1353232015 |
- |
- |
PREDICTED: uncharacterized protein LOC105764482 [Gossypium raimondii] |
| 8 |
Hb_124540_020 |
0.1364829408 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 9 |
Hb_010783_010 |
0.1416999026 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_001817_190 |
0.1418094633 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_001972_020 |
0.1419778186 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_004019_100 |
0.1427321242 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 13 |
Hb_001309_030 |
0.1428712141 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000710_120 |
0.1431311273 |
- |
- |
hypothetical protein JCGZ_01844 [Jatropha curcas] |
| 15 |
Hb_008976_010 |
0.1435279404 |
- |
- |
- |
| 16 |
Hb_009188_010 |
0.1449629092 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 17 |
Hb_011537_090 |
0.1484962103 |
- |
- |
hypothetical protein B456_009G071100 [Gossypium raimondii] |
| 18 |
Hb_097889_010 |
0.1503552061 |
- |
- |
PREDICTED: uncharacterized protein LOC102584707 [Solanum tuberosum] |
| 19 |
Hb_006778_010 |
0.1858977497 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 20 |
Hb_007645_010 |
0.2126286822 |
- |
- |
NdhE, partial (chloroplast) [Podocalyx loranthoides] |