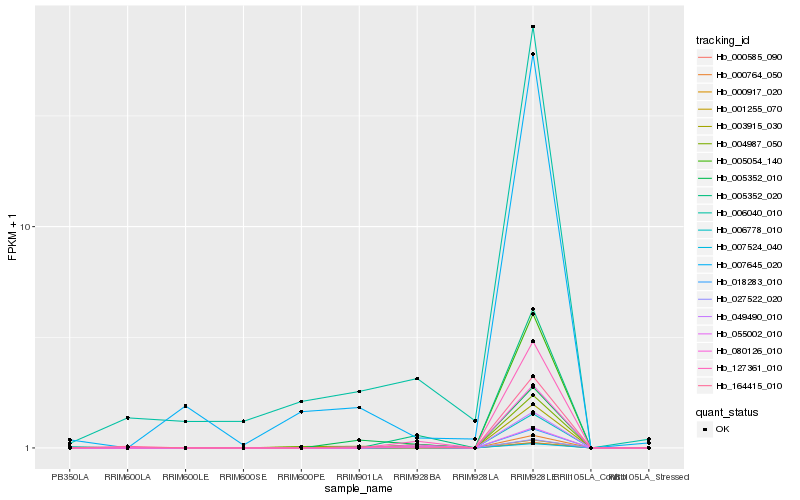
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006778_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 2 |
Hb_027522_020 |
0.1436755042 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_000585_090 |
0.1499163399 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105114407 [Populus euphratica] |
| 4 |
Hb_164415_010 |
0.1582395598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003915_030 |
0.1610703721 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_004987_050 |
0.1632964937 |
- |
- |
- |
| 7 |
Hb_000764_050 |
0.1636610934 |
- |
- |
hypothetical protein JCGZ_09935 [Jatropha curcas] |
| 8 |
Hb_018283_010 |
0.1641610427 |
- |
- |
hypothetical protein VITISV_023498 [Vitis vinifera] |
| 9 |
Hb_006040_010 |
0.1650614257 |
- |
- |
cytochrome c oxidase subunit 3 (mitochondrion) [Hevea brasiliensis] |
| 10 |
Hb_080126_010 |
0.1657657684 |
transcription factor |
TF Family: FAR1 |
hypothetical protein MIMGU_mgv1a025104mg, partial [Erythranthe guttata] |
| 11 |
Hb_127361_010 |
0.1658355416 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 12 |
Hb_055002_010 |
0.167021513 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_049490_010 |
0.1678891686 |
- |
- |
hypothetical protein VITISV_013805 [Vitis vinifera] |
| 14 |
Hb_005054_140 |
0.1694850153 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 15 |
Hb_005352_020 |
0.1698157967 |
- |
- |
- |
| 16 |
Hb_005352_010 |
0.1710489541 |
- |
- |
- |
| 17 |
Hb_007524_040 |
0.1713671326 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC101511096 [Cicer arietinum] |
| 18 |
Hb_000917_020 |
0.1714226396 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 19 |
Hb_007645_020 |
0.1715031417 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit [Medicago truncatula] |
| 20 |
Hb_001255_070 |
0.1716329041 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |