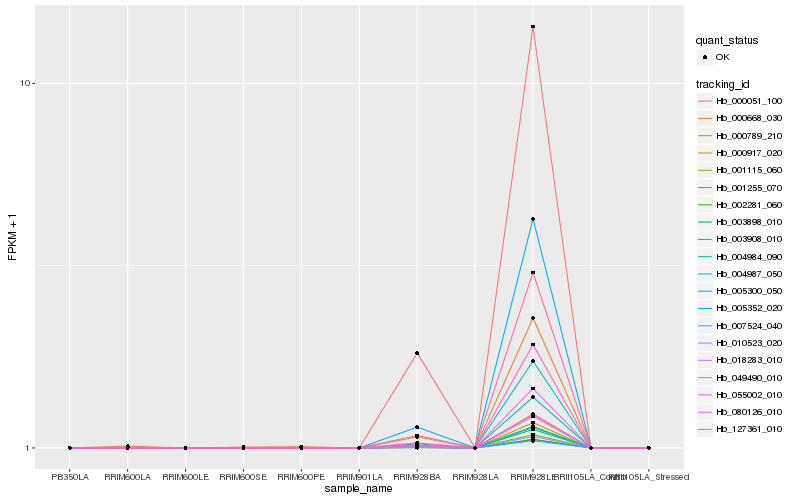
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127361_010 |
0.0 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 2 |
Hb_080126_010 |
0.0003401388 |
transcription factor |
TF Family: FAR1 |
hypothetical protein MIMGU_mgv1a025104mg, partial [Erythranthe guttata] |
| 3 |
Hb_055002_010 |
0.0053778545 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_049490_010 |
0.0089307459 |
- |
- |
hypothetical protein VITISV_013805 [Vitis vinifera] |
| 5 |
Hb_018283_010 |
0.0092760956 |
- |
- |
hypothetical protein VITISV_023498 [Vitis vinifera] |
| 6 |
Hb_005352_020 |
0.0160228565 |
- |
- |
- |
| 7 |
Hb_007524_040 |
0.0211553169 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC101511096 [Cicer arietinum] |
| 8 |
Hb_000917_020 |
0.0213314021 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 9 |
Hb_001255_070 |
0.0219941688 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_003908_010 |
0.0227684529 |
- |
- |
polyprotein [Oryza australiensis] |
| 11 |
Hb_005300_050 |
0.0230608298 |
- |
- |
hypothetical protein VITISV_001308 [Vitis vinifera] |
| 12 |
Hb_003898_010 |
0.0484047376 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 13 |
Hb_010523_020 |
0.0490040966 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_002281_060 |
0.0512434374 |
- |
- |
PREDICTED: uncharacterized protein LOC104877351 [Vitis vinifera] |
| 15 |
Hb_001115_060 |
0.0521476026 |
- |
- |
PREDICTED: uncharacterized protein LOC104602709 [Nelumbo nucifera] |
| 16 |
Hb_000668_030 |
0.0526248896 |
- |
- |
- |
| 17 |
Hb_000789_210 |
0.0543607106 |
- |
- |
PREDICTED: nudix hydrolase 8-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_004987_050 |
0.0550678214 |
- |
- |
- |
| 19 |
Hb_000051_100 |
0.0568036611 |
- |
- |
hypothetical protein MIMGU_mgv1a025286mg [Erythranthe guttata] |
| 20 |
Hb_004984_090 |
0.0591956478 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |