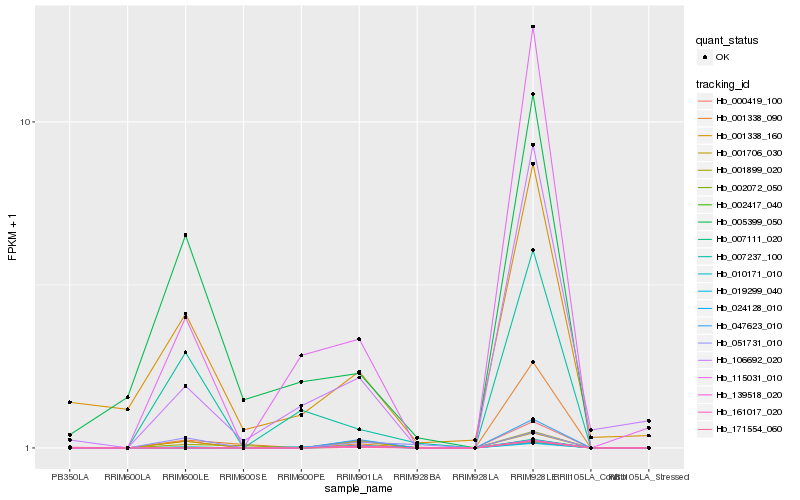
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000419_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_001706_030 |
0.1245638243 |
- |
- |
PREDICTED: uncharacterized protein LOC104880364 [Vitis vinifera] |
| 3 |
Hb_001899_020 |
0.2127361734 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase TPTE2 [Jatropha curcas] |
| 4 |
Hb_002072_050 |
0.2428403684 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_010171_010 |
0.2560620097 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 6 |
Hb_115031_010 |
0.2650403033 |
- |
- |
photosystem II P680 chlorophyll A apoprotein, partial (chloroplast) [Oenothera grandiflora] |
| 7 |
Hb_051731_010 |
0.2720790255 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 8 |
Hb_161017_020 |
0.2835549254 |
- |
- |
hypothetical protein EUTSA_v10012231mg, partial [Eutrema salsugineum] |
| 9 |
Hb_007237_100 |
0.2839110941 |
- |
- |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 10 |
Hb_005399_050 |
0.2856054706 |
- |
- |
DNA-directed RNA polymerase subunit beta [Glycine soja] |
| 11 |
Hb_001338_090 |
0.2856482855 |
- |
- |
hypothetical protein VITISV_009275 [Vitis vinifera] |
| 12 |
Hb_106692_020 |
0.2857770722 |
- |
- |
NADH dehydrogenase subunit F, partial (chloroplast) [Hevea sp. Gillespie 4272] |
| 13 |
Hb_001338_160 |
0.289146077 |
- |
- |
NADH dehydrogenase subunit 5 [Hevea brasiliensis] |
| 14 |
Hb_171554_060 |
0.2918049368 |
- |
- |
PREDICTED: uncharacterized protein LOC102627687 [Citrus sinensis] |
| 15 |
Hb_047623_010 |
0.2918165014 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 16 |
Hb_002417_040 |
0.2918342569 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 17 |
Hb_139518_020 |
0.2918505352 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 18 |
Hb_024128_010 |
0.2918924488 |
- |
- |
PREDICTED: uncharacterized protein LOC105766879 [Gossypium raimondii] |
| 19 |
Hb_019299_040 |
0.2918976211 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor ABI3 [Jatropha curcas] |
| 20 |
Hb_007111_020 |
0.2919708102 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |