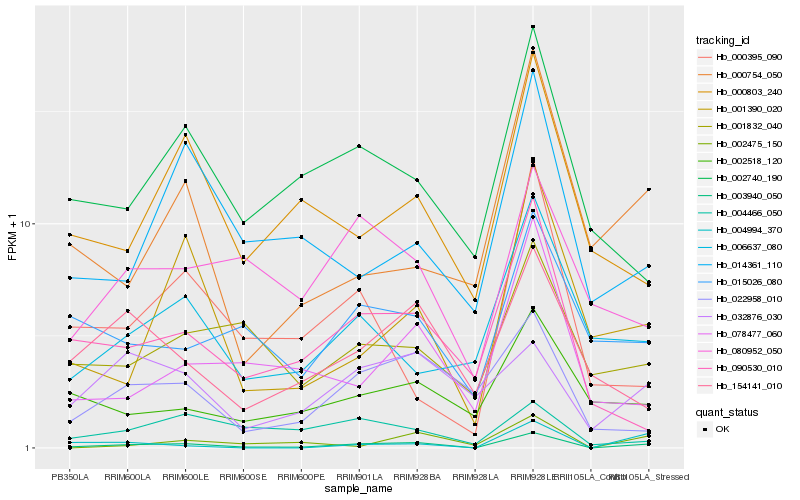
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003940_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 2 |
Hb_001390_020 |
0.2674618922 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_022958_010 |
0.2790982788 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 4 |
Hb_090530_010 |
0.2824573104 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_078477_060 |
0.2841738663 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 6 |
Hb_002475_150 |
0.2863593045 |
- |
- |
PREDICTED: importin subunit beta-3-like [Jatropha curcas] |
| 7 |
Hb_004994_370 |
0.2899041138 |
- |
- |
- |
| 8 |
Hb_006637_080 |
0.2903761158 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002740_190 |
0.2908785217 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_015026_080 |
0.2916069768 |
- |
- |
- |
| 11 |
Hb_154141_010 |
0.2926414718 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 12 |
Hb_001832_040 |
0.29269679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_014361_110 |
0.2929384574 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000803_240 |
0.2938242027 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 15 |
Hb_080952_050 |
0.2956290026 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Nelumbo nucifera] |
| 16 |
Hb_032876_030 |
0.2962610675 |
- |
- |
- |
| 17 |
Hb_002518_120 |
0.2965571371 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |
| 18 |
Hb_000754_050 |
0.297640399 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000395_090 |
0.2977276787 |
- |
- |
PREDICTED: uncharacterized protein LOC105645572 [Jatropha curcas] |
| 20 |
Hb_004466_050 |
0.2980127686 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |