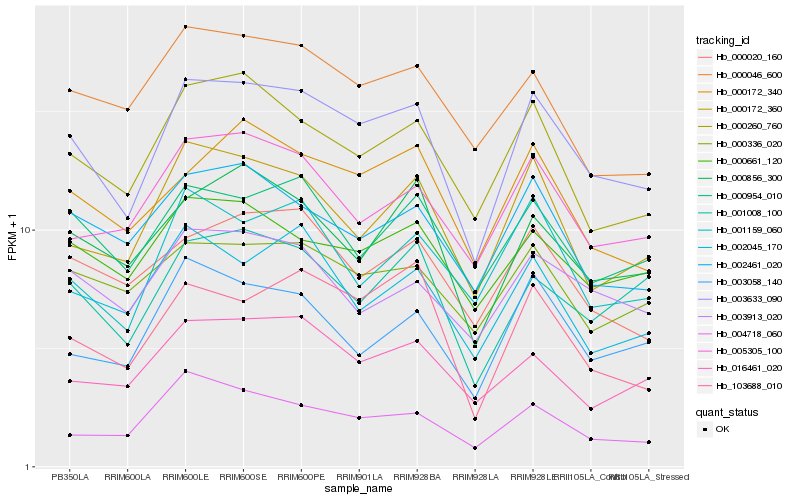
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003633_090 |
0.0 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 2 |
Hb_000260_760 |
0.0884063229 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 3 |
Hb_000172_360 |
0.0930595929 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 4 |
Hb_000172_340 |
0.0951461741 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 5 |
Hb_000661_120 |
0.096050691 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 6 |
Hb_000954_010 |
0.0962079772 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 7 |
Hb_000856_300 |
0.098444777 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 8 |
Hb_002461_020 |
0.0994031736 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000020_160 |
0.0994486192 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 10 |
Hb_001008_100 |
0.0995985921 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 11 |
Hb_005305_100 |
0.1008503245 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000336_020 |
0.1012304428 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 13 |
Hb_016461_020 |
0.101855223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_103688_010 |
0.1030046029 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 15 |
Hb_003913_020 |
0.1031911298 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At5g50170 [Jatropha curcas] |
| 16 |
Hb_003058_140 |
0.1035866547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000046_600 |
0.1052378093 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 18 |
Hb_001159_060 |
0.1054028661 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 19 |
Hb_002045_170 |
0.1058279387 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 20 |
Hb_004718_060 |
0.1064823291 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |