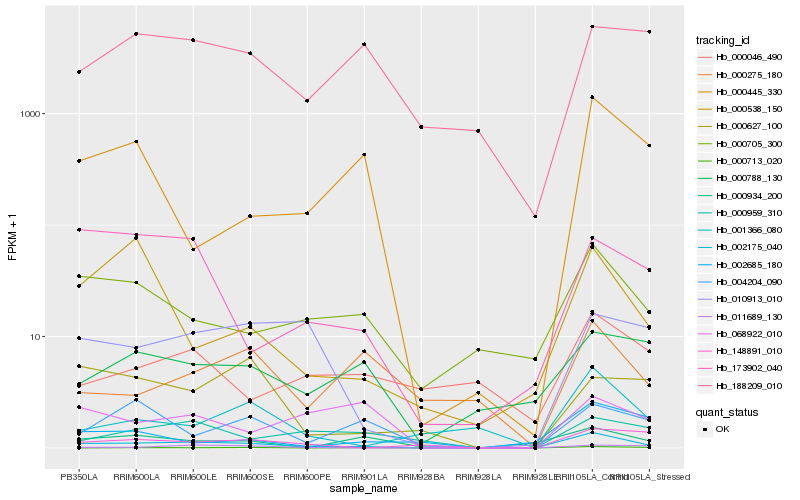
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002175_040 |
0.0 |
- |
- |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 2 |
Hb_000705_300 |
0.270875372 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 3 |
Hb_001366_080 |
0.28607865 |
transcription factor |
TF Family: NAC |
NAC transcription factor 068 [Jatropha curcas] |
| 4 |
Hb_011689_130 |
0.2943296136 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_010913_010 |
0.2948460046 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_148891_010 |
0.2954980092 |
- |
- |
- |
| 7 |
Hb_000445_330 |
0.2984756024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000934_200 |
0.3052862853 |
- |
- |
PREDICTED: uncharacterized protein LOC105633952 [Jatropha curcas] |
| 9 |
Hb_173902_040 |
0.3053257285 |
- |
- |
- |
| 10 |
Hb_000788_130 |
0.3078750579 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 11 |
Hb_000959_310 |
0.3114324367 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 12 |
Hb_188209_010 |
0.3122905851 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_004204_090 |
0.3170204308 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 14 |
Hb_068922_010 |
0.3200740059 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 15 |
Hb_000275_180 |
0.320153349 |
- |
- |
- |
| 16 |
Hb_000538_150 |
0.3204317262 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 17 |
Hb_000627_100 |
0.3211361342 |
- |
- |
- |
| 18 |
Hb_000046_490 |
0.3251397648 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 19 |
Hb_000713_020 |
0.3261727902 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_002685_180 |
0.329184543 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |