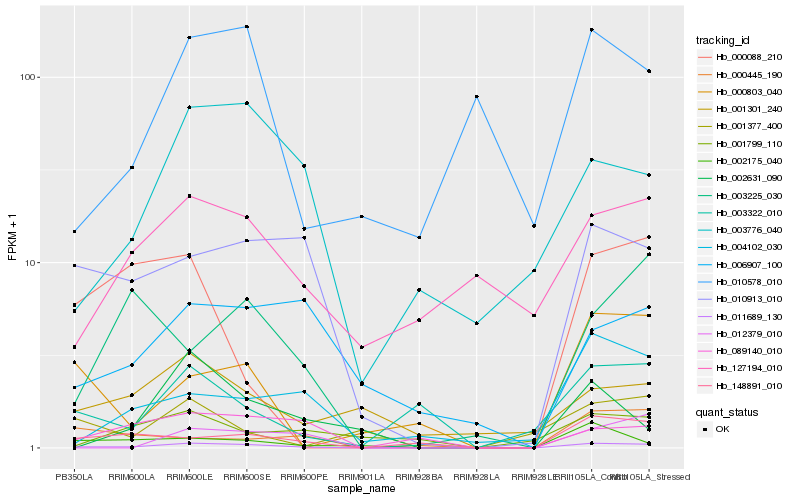
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011689_130 |
0.0 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_010913_010 |
0.2487744227 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 3 |
Hb_148891_010 |
0.2660125673 |
- |
- |
- |
| 4 |
Hb_003322_010 |
0.2887585402 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003776_040 |
0.290258979 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 6 |
Hb_002175_040 |
0.2943296136 |
- |
- |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 7 |
Hb_004102_030 |
0.2983714188 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 8 |
Hb_089140_010 |
0.3007228999 |
- |
- |
- |
| 9 |
Hb_000803_040 |
0.3059187454 |
- |
- |
- |
| 10 |
Hb_006907_100 |
0.3079390939 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 11 |
Hb_010578_010 |
0.3079871637 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 12 |
Hb_012379_010 |
0.3085935459 |
- |
- |
- |
| 13 |
Hb_001301_240 |
0.314657011 |
- |
- |
- |
| 14 |
Hb_127194_010 |
0.315955295 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 15 |
Hb_003225_030 |
0.3164211961 |
- |
- |
Pectinesterase-4 precursor, putative [Ricinus communis] |
| 16 |
Hb_002631_090 |
0.3218393577 |
- |
- |
- |
| 17 |
Hb_000445_190 |
0.3219545685 |
- |
- |
- |
| 18 |
Hb_000088_210 |
0.3251544148 |
- |
- |
hypothetical protein POPTR_0013s11310g [Populus trichocarpa] |
| 19 |
Hb_001377_400 |
0.3290006533 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 20 |
Hb_001799_110 |
0.3293180954 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |