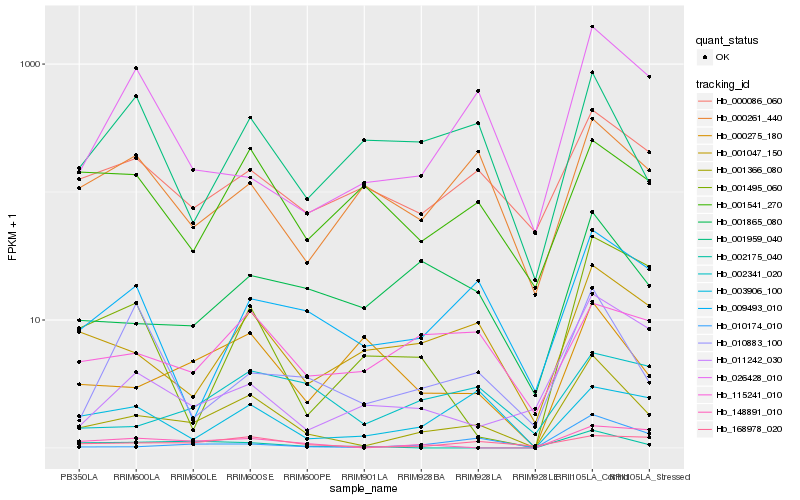
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_080 |
0.0 |
transcription factor |
TF Family: NAC |
NAC transcription factor 068 [Jatropha curcas] |
| 2 |
Hb_001047_150 |
0.2534081301 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 3 |
Hb_168978_020 |
0.2694112685 |
- |
- |
hypothetical protein POPTR_0002s10960g [Populus trichocarpa] |
| 4 |
Hb_001865_080 |
0.2726132748 |
- |
- |
enoyl-CoA hydratase/isomerase family protein [Populus trichocarpa] |
| 5 |
Hb_000086_060 |
0.2763372015 |
- |
- |
PREDICTED: TOM1-like protein 2 isoform X2 [Populus euphratica] |
| 6 |
Hb_148891_010 |
0.278183642 |
- |
- |
- |
| 7 |
Hb_009493_010 |
0.2796879723 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 8 |
Hb_010174_010 |
0.285650473 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 9 |
Hb_002175_040 |
0.28607865 |
- |
- |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 10 |
Hb_000275_180 |
0.2871565118 |
- |
- |
- |
| 11 |
Hb_115241_010 |
0.2933731589 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 12 |
Hb_011242_030 |
0.2953138378 |
- |
- |
- |
| 13 |
Hb_000261_440 |
0.3008564653 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_026428_010 |
0.3046776132 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 15 |
Hb_003906_100 |
0.3051342337 |
- |
- |
hypothetical protein JCGZ_26174 [Jatropha curcas] |
| 16 |
Hb_002341_020 |
0.3064787044 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 17 |
Hb_001959_040 |
0.3068330445 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001495_060 |
0.3076390023 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 19 |
Hb_010883_100 |
0.3078849159 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 20 |
Hb_001541_270 |
0.3086197875 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |