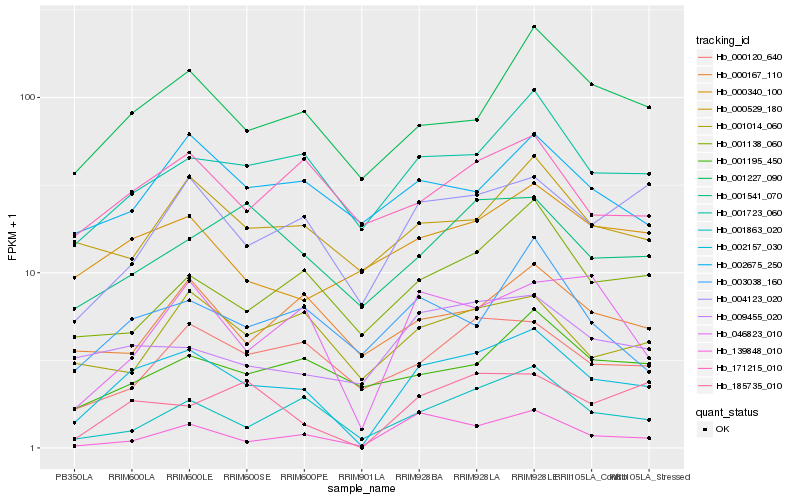
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001723_060 |
0.1705010033 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 3 |
Hb_001227_090 |
0.1746227744 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 4 |
Hb_004123_020 |
0.1763426771 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_001863_020 |
0.1829996266 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_185735_010 |
0.1841560321 |
- |
- |
Heat shock factor protein HSF8, putative [Ricinus communis] |
| 7 |
Hb_046823_010 |
0.1845643919 |
- |
- |
hypothetical protein CICLE_v100021301mg, partial [Citrus clementina] |
| 8 |
Hb_009455_020 |
0.1898239774 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 9 |
Hb_000120_640 |
0.192339229 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000529_180 |
0.1923932733 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_139848_010 |
0.1942659696 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |
| 12 |
Hb_000340_100 |
0.1961645365 |
- |
- |
PREDICTED: protein PALE CRESS, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001138_060 |
0.1965929346 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 14 |
Hb_171215_010 |
0.1976547006 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 15 |
Hb_003038_160 |
0.1987994074 |
- |
- |
auxilin, putative [Ricinus communis] |
| 16 |
Hb_002675_250 |
0.1990349729 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 17 |
Hb_001014_060 |
0.1992350312 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 18 |
Hb_001541_070 |
0.199447754 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 19 |
Hb_001195_450 |
0.2019832169 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 20 |
Hb_000167_110 |
0.2021446573 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |