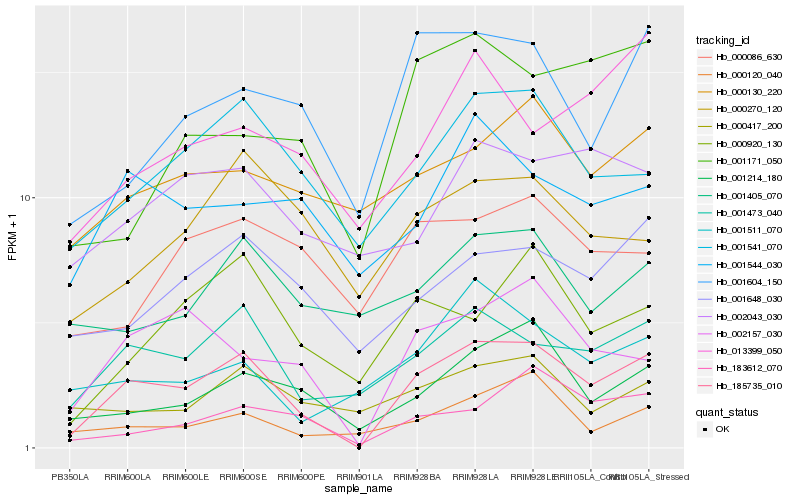
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185735_010 |
0.0 |
- |
- |
Heat shock factor protein HSF8, putative [Ricinus communis] |
| 2 |
Hb_001473_040 |
0.1718080203 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_002157_030 |
0.1841560321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001541_070 |
0.1954762511 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 5 |
Hb_001214_180 |
0.1956125094 |
- |
- |
- |
| 6 |
Hb_001604_150 |
0.2019925964 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Nelumbo nucifera] |
| 7 |
Hb_183612_070 |
0.2019981579 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000920_130 |
0.2032733482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001648_030 |
0.206208096 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000120_040 |
0.2134606889 |
- |
- |
- |
| 11 |
Hb_001544_030 |
0.2186018922 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1C [Jatropha curcas] |
| 12 |
Hb_000270_120 |
0.2194574741 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 13 |
Hb_001171_050 |
0.2222297434 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 14 |
Hb_001511_070 |
0.2223158361 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 15 |
Hb_000417_200 |
0.2223499831 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002043_030 |
0.2225084777 |
- |
- |
hypothetical protein RCOM_1486170 [Ricinus communis] |
| 17 |
Hb_000130_220 |
0.2228105477 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_001405_070 |
0.2248347009 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 19 |
Hb_000086_630 |
0.2251738869 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_013399_050 |
0.2262217314 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 1-like isoform X2 [Jatropha curcas] |