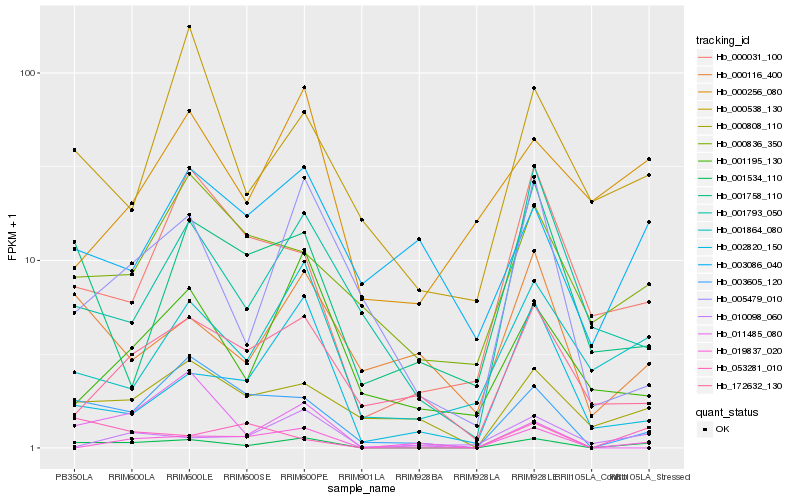
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001534_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003605_120 |
0.2574568324 |
- |
- |
hypothetical protein POPTR_0013s12060g [Populus trichocarpa] |
| 3 |
Hb_001864_080 |
0.2858968305 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |
| 4 |
Hb_000808_110 |
0.2933711852 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X4 [Jatropha curcas] |
| 5 |
Hb_005479_010 |
0.3007209364 |
- |
- |
PREDICTED: uncharacterized protein LOC105111002 [Populus euphratica] |
| 6 |
Hb_000116_400 |
0.3037617681 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_053281_010 |
0.3155114537 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_002820_150 |
0.3167618069 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000031_100 |
0.3192712941 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 10 |
Hb_001793_050 |
0.3205359547 |
- |
- |
PREDICTED: uncharacterized protein LOC105632663 [Jatropha curcas] |
| 11 |
Hb_172632_130 |
0.3234057238 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 12 |
Hb_000538_130 |
0.3257465195 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 13 |
Hb_001195_130 |
0.326178751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_019837_020 |
0.3266483938 |
- |
- |
- |
| 15 |
Hb_000256_080 |
0.3267911808 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 16 |
Hb_011485_080 |
0.3276947734 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 17 |
Hb_010098_060 |
0.3286489366 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g17580 [Jatropha curcas] |
| 18 |
Hb_003086_040 |
0.329155065 |
- |
- |
PREDICTED: uncharacterized protein LOC105635780 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001758_110 |
0.3294282268 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 20 |
Hb_000836_350 |
0.3294997951 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |