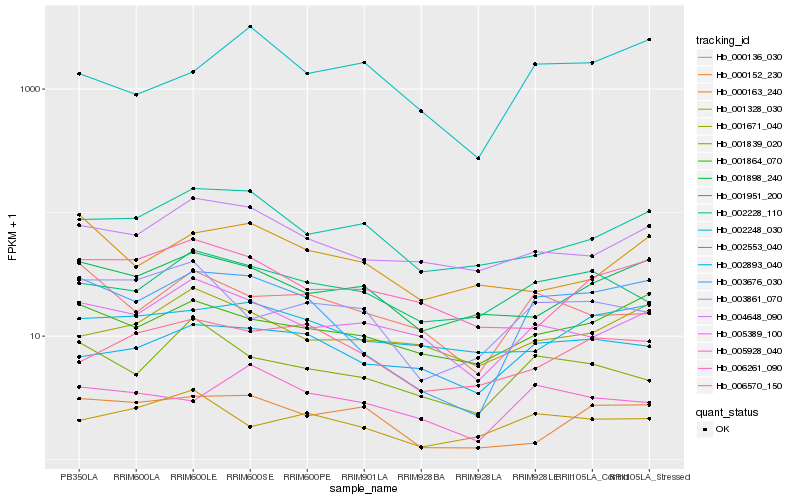
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003676_030 |
0.0 |
- |
- |
PREDICTED: cyclin-L1-1-like [Populus euphratica] |
| 2 |
Hb_001864_070 |
0.1709551633 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |
| 3 |
Hb_002893_040 |
0.1816741433 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000136_030 |
0.1846026011 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 5 |
Hb_001839_020 |
0.1847358076 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 6 |
Hb_005389_100 |
0.1856630948 |
- |
- |
PREDICTED: methyltransferase-like protein 5 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004648_090 |
0.1863730796 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 8 |
Hb_001898_240 |
0.1886414949 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 9 |
Hb_005928_040 |
0.1911742867 |
- |
- |
phosphoinositide phosphatase family protein [Populus trichocarpa] |
| 10 |
Hb_003861_070 |
0.1955217059 |
- |
- |
PREDICTED: uncharacterized protein LOC105650781 [Jatropha curcas] |
| 11 |
Hb_000152_230 |
0.1962190714 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 12 |
Hb_002553_040 |
0.1975368803 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 13 |
Hb_002228_110 |
0.1988358397 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 14 |
Hb_006261_090 |
0.1999547267 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 15 |
Hb_001951_200 |
0.2014914168 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_006570_150 |
0.2017644826 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001328_030 |
0.201884136 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 18 |
Hb_002248_030 |
0.2041342965 |
- |
- |
metallothionein-like protein [Hevea brasiliensis] |
| 19 |
Hb_001671_040 |
0.2051843481 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000163_240 |
0.2057476173 |
- |
- |
PREDICTED: protein OPI10 homolog [Jatropha curcas] |