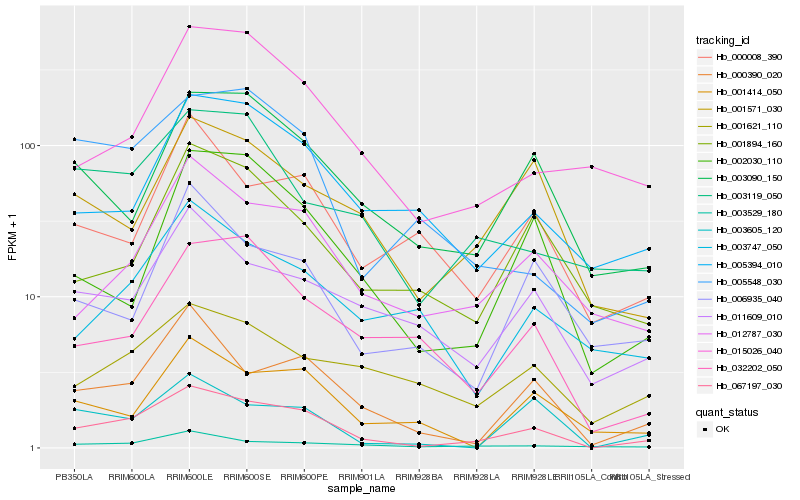
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_067197_030 |
0.0 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000390_020 |
0.1675546681 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 3 |
Hb_012787_030 |
0.1887820162 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 4 |
Hb_003090_150 |
0.194938855 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 5 |
Hb_003529_180 |
0.1955965526 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 6 |
Hb_002030_110 |
0.1991663408 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 7 |
Hb_011609_010 |
0.2013292688 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 8 |
Hb_001571_030 |
0.2043133924 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 9 |
Hb_015026_040 |
0.2054266185 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 10 |
Hb_005394_010 |
0.2054850167 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 11 |
Hb_003605_120 |
0.2060640222 |
- |
- |
hypothetical protein POPTR_0013s12060g [Populus trichocarpa] |
| 12 |
Hb_003119_050 |
0.2068816032 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001894_160 |
0.2073990558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001621_110 |
0.209672832 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |
| 15 |
Hb_005548_030 |
0.2107494422 |
- |
- |
hypothetical protein JCGZ_05606 [Jatropha curcas] |
| 16 |
Hb_032202_050 |
0.2110577528 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003747_050 |
0.2143615687 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 18 |
Hb_006935_040 |
0.2148814546 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 19 |
Hb_001414_050 |
0.215286591 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 20 |
Hb_000008_390 |
0.2163227063 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |