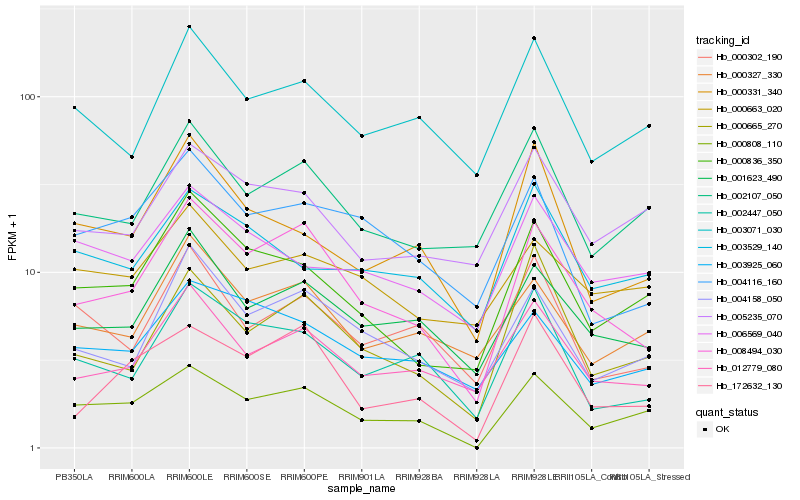
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000808_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X4 [Jatropha curcas] |
| 2 |
Hb_000836_350 |
0.1414853747 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |
| 3 |
Hb_008494_030 |
0.1424855774 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_006569_040 |
0.1578277069 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 5 |
Hb_002107_050 |
0.1588641348 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 6 |
Hb_001623_490 |
0.1589270982 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000331_340 |
0.1594545043 |
- |
- |
PREDICTED: uncharacterized protein LOC105640234 [Jatropha curcas] |
| 8 |
Hb_003071_030 |
0.1606542798 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000327_330 |
0.1642699122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005235_070 |
0.1644014438 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003529_140 |
0.1647591855 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002447_050 |
0.1656268955 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000665_270 |
0.1656560557 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 14 |
Hb_003925_060 |
0.1658235255 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 15 |
Hb_172632_130 |
0.1663863746 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 16 |
Hb_000302_190 |
0.1668245437 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 17 |
Hb_004116_160 |
0.1679545655 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004158_050 |
0.1686071476 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 19 |
Hb_000663_020 |
0.1689160032 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_012779_080 |
0.1697416396 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |