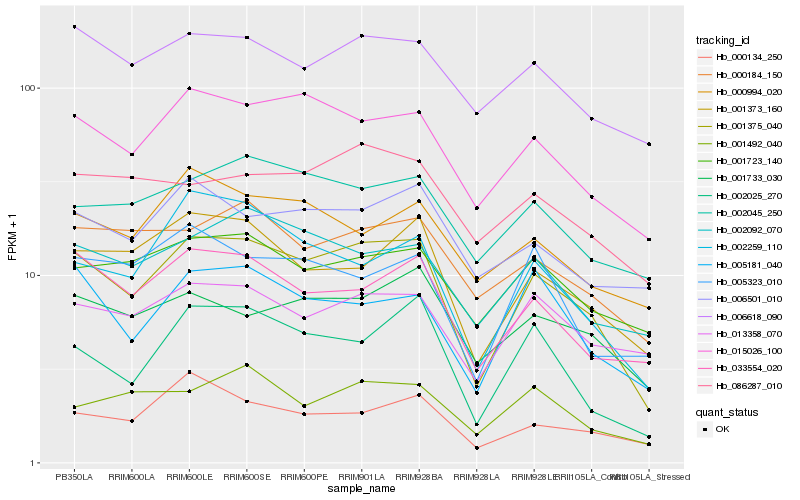
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001375_040 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_015026_100 |
0.1033792694 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 3 |
Hb_033554_020 |
0.1141906082 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 4 |
Hb_001373_160 |
0.1156675776 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 5 |
Hb_002025_270 |
0.1178950703 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002259_110 |
0.123600345 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 7 |
Hb_005181_040 |
0.1247122878 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 8 |
Hb_002092_070 |
0.1255077774 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 9 |
Hb_000184_150 |
0.1267942212 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 10 |
Hb_006618_090 |
0.1274463814 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 11 |
Hb_000134_250 |
0.1288641332 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_001733_030 |
0.1292372847 |
- |
- |
PREDICTED: uncharacterized protein LOC100261386 isoform X2 [Vitis vinifera] |
| 13 |
Hb_002045_250 |
0.1301774043 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 14 |
Hb_013358_070 |
0.1316173368 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_001723_140 |
0.132531083 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_006501_010 |
0.1353594733 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A isoform X1 [Populus euphratica] |
| 17 |
Hb_000994_020 |
0.135978535 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_086287_010 |
0.1361539235 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001492_040 |
0.1361733358 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005323_010 |
0.1374970021 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |