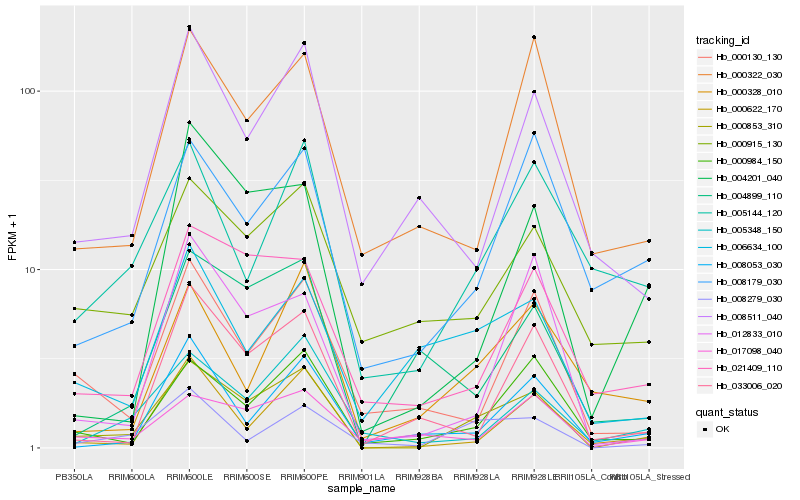
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_310 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Populus euphratica] |
| 2 |
Hb_000984_150 |
0.2040724199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000622_170 |
0.2103120641 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 4 |
Hb_021409_110 |
0.2170050095 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_008279_030 |
0.2233392834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000915_130 |
0.223946806 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 7 |
Hb_005348_150 |
0.2270957331 |
- |
- |
PREDICTED: actin-100-like [Jatropha curcas] |
| 8 |
Hb_017098_040 |
0.2294781928 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 9 |
Hb_004201_040 |
0.2321407359 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 10 |
Hb_008179_030 |
0.2331983027 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 8 [Jatropha curcas] |
| 11 |
Hb_033006_020 |
0.2343590761 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 12 |
Hb_008511_040 |
0.2345701208 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_000130_130 |
0.2349190064 |
- |
- |
hypothetical protein POPTR_0001s40330g [Populus trichocarpa] |
| 14 |
Hb_000322_030 |
0.2361704206 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 15 |
Hb_005144_120 |
0.237381231 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 16 |
Hb_004899_110 |
0.2397645123 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 17 |
Hb_012833_010 |
0.2399309111 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006634_100 |
0.2408396406 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000328_010 |
0.2418176806 |
- |
- |
PREDICTED: QWRF motif-containing protein 8-like [Jatropha curcas] |
| 20 |
Hb_008053_030 |
0.2420907056 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |