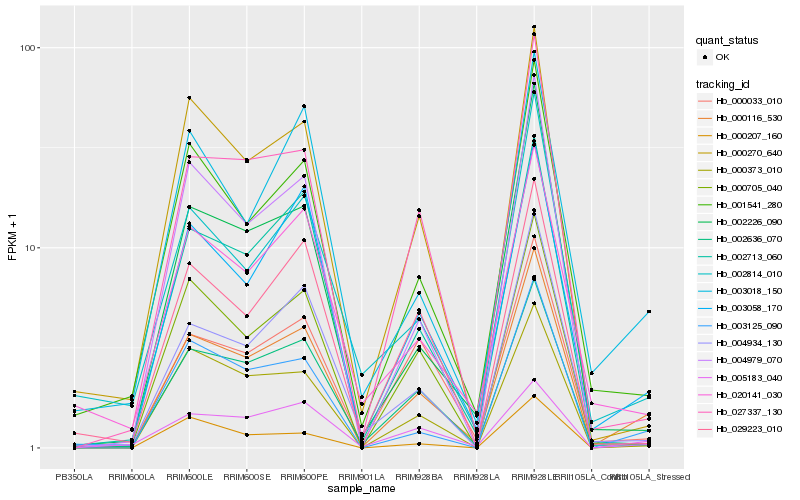
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_530 |
0.0 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 3; Short=HMG-CoA reductase 3 [Hevea brasiliensis] |
| 2 |
Hb_003058_170 |
0.1126477563 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003125_090 |
0.1165772178 |
- |
- |
- |
| 4 |
Hb_002636_070 |
0.1183525421 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 5 |
Hb_001541_280 |
0.1208721611 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 6 |
Hb_027337_130 |
0.1230523573 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 7 |
Hb_004934_130 |
0.1303065508 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 8 |
Hb_000270_640 |
0.1316445554 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 9 |
Hb_002713_060 |
0.1339429213 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 10 |
Hb_000705_040 |
0.1349938266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000033_010 |
0.1366628688 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 12 |
Hb_002226_090 |
0.1376350711 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 13 |
Hb_000207_160 |
0.1378131623 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Populus euphratica] |
| 14 |
Hb_003018_150 |
0.1385072761 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 15 |
Hb_020141_030 |
0.1391996081 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_000373_010 |
0.1415271496 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_029223_010 |
0.1422655446 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 18 |
Hb_002814_010 |
0.1430145449 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005183_040 |
0.1449391364 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 20 |
Hb_004979_070 |
0.1451073267 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |