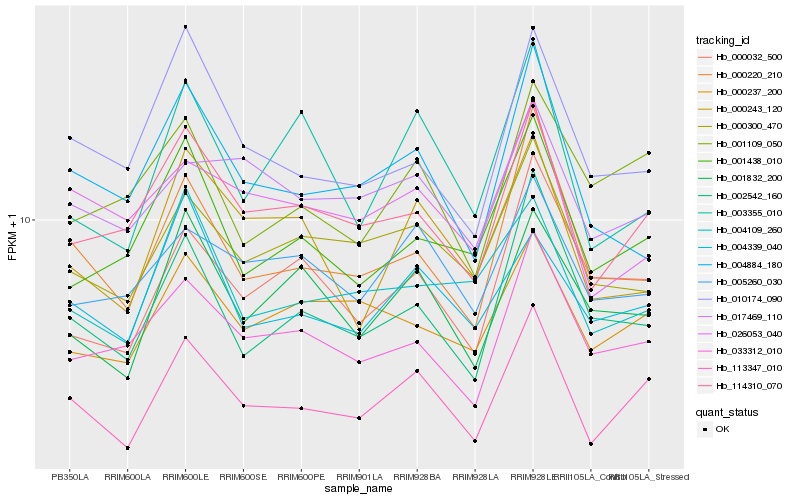
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_113347_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g009336mg [Citrus sinensis] |
| 2 |
Hb_114310_070 |
0.0930739875 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 3 |
Hb_000220_210 |
0.1011689781 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002542_160 |
0.1087058651 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_033312_010 |
0.1118202909 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_001832_200 |
0.1118222375 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000300_470 |
0.1131015692 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 8 |
Hb_000237_200 |
0.1150659843 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 9 |
Hb_004339_040 |
0.1165491349 |
- |
- |
hypothetical protein POPTR_0162s00290g, partial [Populus trichocarpa] |
| 10 |
Hb_001438_010 |
0.1181477659 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000032_500 |
0.1183684276 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 12 |
Hb_010174_090 |
0.1185999955 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005260_030 |
0.1191738726 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 14 |
Hb_001109_050 |
0.1198226313 |
- |
- |
PREDICTED: ankyrin-1 [Jatropha curcas] |
| 15 |
Hb_000243_120 |
0.1201221859 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_004884_180 |
0.1215488153 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003355_010 |
0.1221726161 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 18 |
Hb_017469_110 |
0.1228509255 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 19 |
Hb_004109_260 |
0.1240542078 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 20 |
Hb_026053_040 |
0.1240578704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |