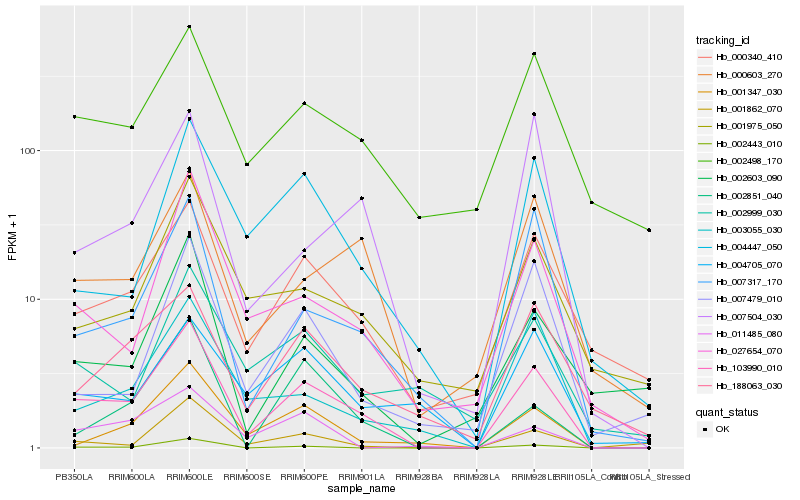
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103990_010 |
0.0 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 2 |
Hb_000340_410 |
0.1960637631 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_027654_070 |
0.1972761748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007317_170 |
0.2018043895 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 5 |
Hb_000603_270 |
0.2085885901 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 6 |
Hb_001347_030 |
0.217535984 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_004447_050 |
0.2191495994 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_188063_030 |
0.2201136969 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 9 |
Hb_002498_170 |
0.2217334585 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002443_010 |
0.2225012925 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 11 |
Hb_003055_030 |
0.2235468416 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 12 |
Hb_001862_070 |
0.2239821088 |
- |
- |
hypothetical protein RCOM_1155060 [Ricinus communis] |
| 13 |
Hb_002603_090 |
0.2245082588 |
- |
- |
PREDICTED: GDSL esterase/lipase CPRD49-like isoform X2 [Populus euphratica] |
| 14 |
Hb_001975_050 |
0.2261407357 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 15 |
Hb_007504_030 |
0.2320704435 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 16 |
Hb_007479_010 |
0.233598427 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 17 |
Hb_002851_040 |
0.2350523472 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 18 |
Hb_004705_070 |
0.236911656 |
- |
- |
PREDICTED: uncharacterized protein LOC105647312 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011485_080 |
0.2369853074 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002999_030 |
0.2377545008 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |