| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
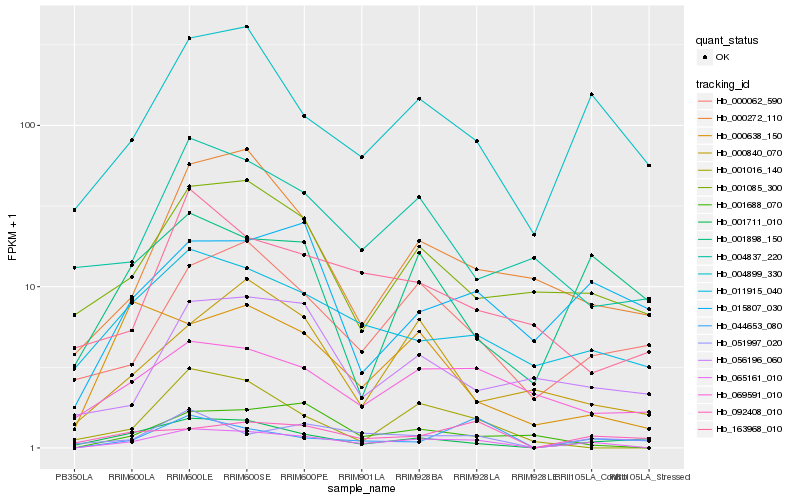
Hb_065161_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004899_330 |
0.2333017547 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 3 |
Hb_011915_040 |
0.2540957742 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 4 |
Hb_000062_590 |
0.2546953049 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_001688_070 |
0.2564116243 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X1 [Jatropha curcas] |
| 6 |
Hb_051997_020 |
0.258824571 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 7 |
Hb_001711_010 |
0.262158005 |
- |
- |
- |
| 8 |
Hb_001085_300 |
0.2652048718 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 9 |
Hb_000272_110 |
0.2675782822 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001016_140 |
0.2686993022 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 11 |
Hb_069591_010 |
0.2724155545 |
- |
- |
- |
| 12 |
Hb_092408_010 |
0.2724207476 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_044653_080 |
0.2780056274 |
- |
- |
Uncharacterized protein TCM_045144 [Theobroma cacao] |
| 14 |
Hb_015807_030 |
0.2790986021 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 15 |
Hb_000638_150 |
0.2806818504 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 16 |
Hb_001898_150 |
0.2828277311 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 17 |
Hb_004837_220 |
0.2830127477 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 18 |
Hb_163968_010 |
0.2831346986 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 19 |
Hb_056196_060 |
0.2845677368 |
- |
- |
hexokinase [Manihot esculenta] |
| 20 |
Hb_000840_070 |
0.2850373701 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |