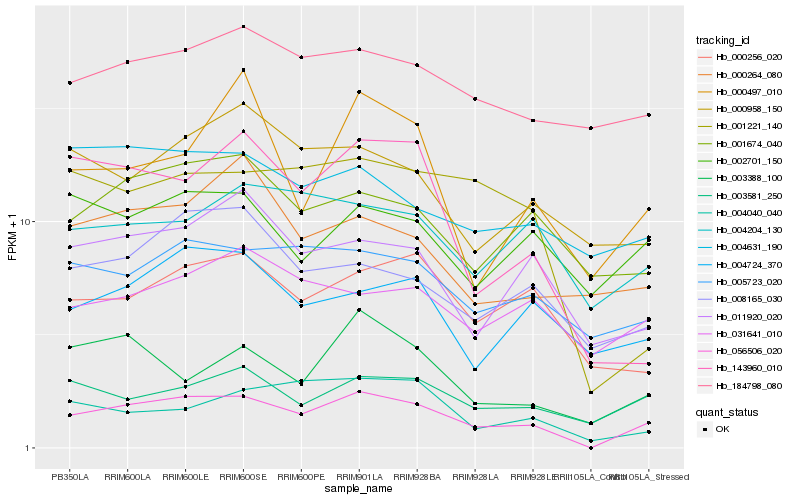
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_056506_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647588 [Jatropha curcas] |
| 2 |
Hb_003581_250 |
0.1684156276 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 3 |
Hb_005723_020 |
0.1685081558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011920_020 |
0.1701764506 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 5 |
Hb_008165_030 |
0.171596027 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 6 |
Hb_001674_040 |
0.173250382 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 7 |
Hb_000256_020 |
0.1736302835 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 8 |
Hb_003388_100 |
0.1742572046 |
- |
- |
16S rRNA processing protein RimM [Psychrobacter phenylpyruvicus] |
| 9 |
Hb_004724_370 |
0.1762064995 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 10 |
Hb_143960_010 |
0.1783577691 |
- |
- |
- |
| 11 |
Hb_184798_080 |
0.1787077904 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 12 |
Hb_004204_130 |
0.1789042893 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 13 |
Hb_001221_140 |
0.1792279449 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_002701_150 |
0.1802694719 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_031641_010 |
0.1808324257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000497_010 |
0.1817981501 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR2 [Jatropha curcas] |
| 17 |
Hb_004040_040 |
0.1824123018 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 18 |
Hb_000958_150 |
0.1831600656 |
- |
- |
Actin, putative [Ricinus communis] |
| 19 |
Hb_004631_190 |
0.1848460238 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 20 |
Hb_000264_080 |
0.1851808564 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |