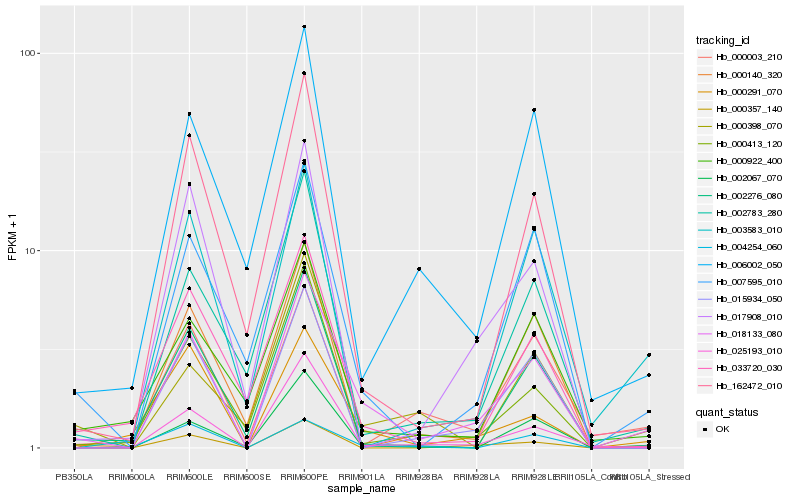
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018133_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_007595_010 |
0.191330423 |
- |
- |
Photosystem II reaction center protein K [Medicago truncatula] |
| 3 |
Hb_000291_070 |
0.2034347867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_025193_010 |
0.2089827082 |
- |
- |
PREDICTED: indole-3-acetic acid-amido synthetase GH3.17-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_015934_050 |
0.20945628 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 6 |
Hb_162472_010 |
0.2107600273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002783_280 |
0.2133812707 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 8 |
Hb_002276_080 |
0.21982541 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 9 |
Hb_006002_050 |
0.2213048578 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 10 |
Hb_000413_120 |
0.2218849772 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 11 |
Hb_033720_030 |
0.2230795244 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_002067_070 |
0.2250332952 |
- |
- |
hypothetical protein JCGZ_19254 [Jatropha curcas] |
| 13 |
Hb_000922_400 |
0.2255051674 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 14 |
Hb_000357_140 |
0.2256598561 |
- |
- |
hypothetical protein JCGZ_15089 [Jatropha curcas] |
| 15 |
Hb_004254_060 |
0.2268174524 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 16 |
Hb_000140_320 |
0.228295824 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 17 |
Hb_017908_010 |
0.2283145609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000398_070 |
0.2284165222 |
- |
- |
hypothetical protein VITISV_011880 [Vitis vinifera] |
| 19 |
Hb_000003_210 |
0.2289224344 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 20 |
Hb_003583_010 |
0.23043998 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |