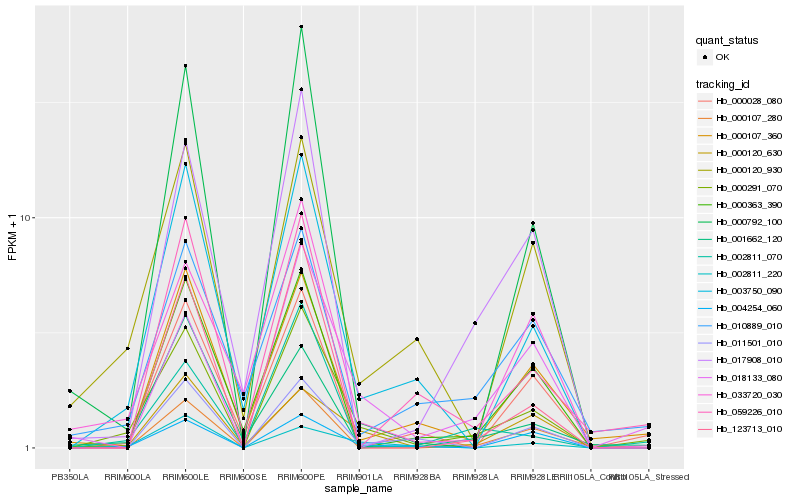
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003750_090 |
0.1847890789 |
- |
- |
BnaC02g34020D [Brassica napus] |
| 3 |
Hb_004254_060 |
0.2008674079 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 4 |
Hb_018133_080 |
0.2034347867 |
- |
- |
- |
| 5 |
Hb_000120_930 |
0.2088715395 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000120_630 |
0.2099598314 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 7 |
Hb_000107_360 |
0.2122285189 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 8 |
Hb_000792_100 |
0.2126906494 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 9 |
Hb_002811_070 |
0.2149071934 |
- |
- |
Cytidine/deoxycytidylate deaminase family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_017908_010 |
0.216242027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011501_010 |
0.2174778015 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG5-like [Jatropha curcas] |
| 12 |
Hb_123713_010 |
0.2185727813 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_010889_010 |
0.2194517998 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 14 |
Hb_000028_080 |
0.2196601025 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 15 |
Hb_000363_390 |
0.2197932111 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_033720_030 |
0.221940108 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_002811_220 |
0.2221749297 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_001662_120 |
0.2236007522 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 19 |
Hb_000107_280 |
0.2237583344 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_059226_010 |
0.2255652594 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |