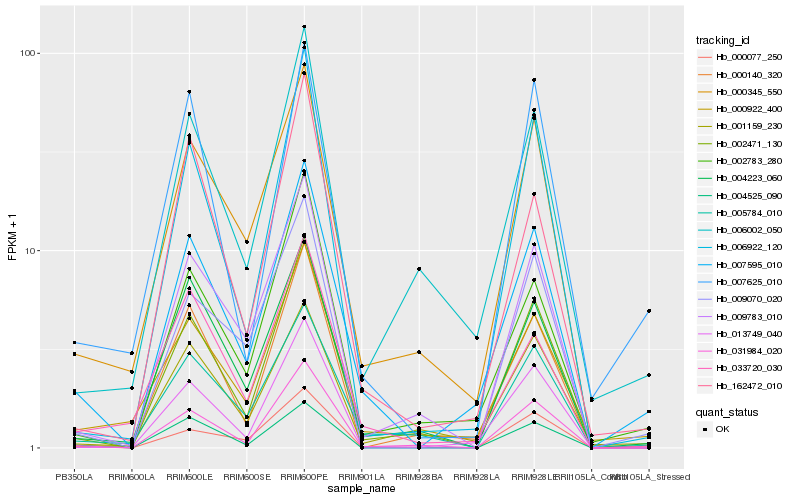
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007595_010 |
0.0 |
- |
- |
Photosystem II reaction center protein K [Medicago truncatula] |
| 2 |
Hb_000922_400 |
0.134674095 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 3 |
Hb_013749_040 |
0.1390766796 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PXL1 [Jatropha curcas] |
| 4 |
Hb_002783_280 |
0.1403777986 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 5 |
Hb_000345_550 |
0.1438291144 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 6 |
Hb_005784_010 |
0.1459924616 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 7 |
Hb_002471_130 |
0.1493094708 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 8 |
Hb_031984_020 |
0.1526777287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004223_060 |
0.1548721513 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 10 |
Hb_001159_230 |
0.1560518692 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 11 |
Hb_006002_050 |
0.1570284208 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 12 |
Hb_006922_120 |
0.1573953354 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 13 |
Hb_000140_320 |
0.1584900739 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 14 |
Hb_033720_030 |
0.1591818228 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_009070_020 |
0.1601320818 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 16 |
Hb_004525_090 |
0.1601566568 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000077_250 |
0.1612905857 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 18 |
Hb_009783_010 |
0.1616640389 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 19 |
Hb_007625_010 |
0.1619319554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_162472_010 |
0.1628804836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |