| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
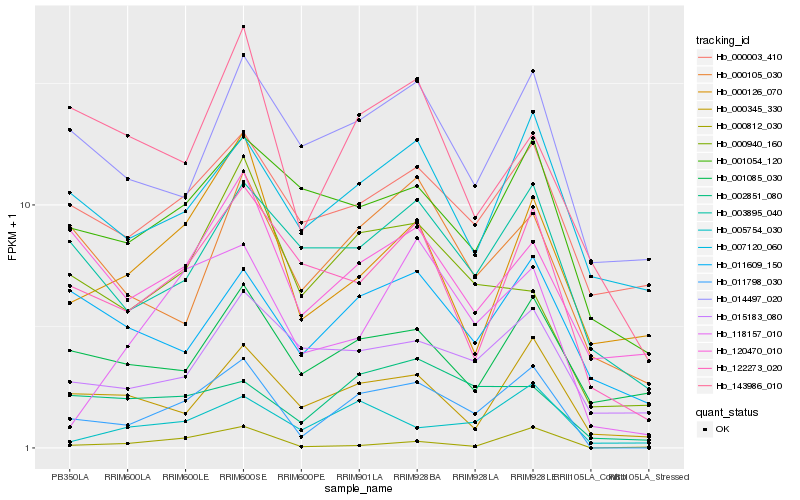
Hb_011798_030 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Vitis vinifera] |
| 2 |
Hb_122273_020 |
0.2082518006 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP2 [Jatropha curcas] |
| 3 |
Hb_118157_010 |
0.2124116319 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 4 |
Hb_000345_330 |
0.214533111 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance-like protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_003895_040 |
0.2189044137 |
- |
- |
hypothetical protein CISIN_1g0006512mg, partial [Citrus sinensis] |
| 6 |
Hb_000940_160 |
0.2234028981 |
- |
- |
hypothetical protein VITISV_017596 [Vitis vinifera] |
| 7 |
Hb_005754_030 |
0.2256879719 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 8 |
Hb_002851_080 |
0.2265004971 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 9 |
Hb_000105_030 |
0.2279726511 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 10 |
Hb_143986_010 |
0.2297066349 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 11 |
Hb_014497_020 |
0.2297907984 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 12 |
Hb_007120_060 |
0.2299691849 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 13 |
Hb_001054_120 |
0.2311071275 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 14 |
Hb_011609_150 |
0.2331037594 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |
| 15 |
Hb_000126_070 |
0.2336864636 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Gossypium arboreum] |
| 16 |
Hb_015183_080 |
0.2336989977 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 17 |
Hb_000003_410 |
0.2339297979 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 18 |
Hb_000812_030 |
0.2344841376 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 19 |
Hb_120470_010 |
0.2345060317 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 20 |
Hb_001085_030 |
0.2348329365 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |