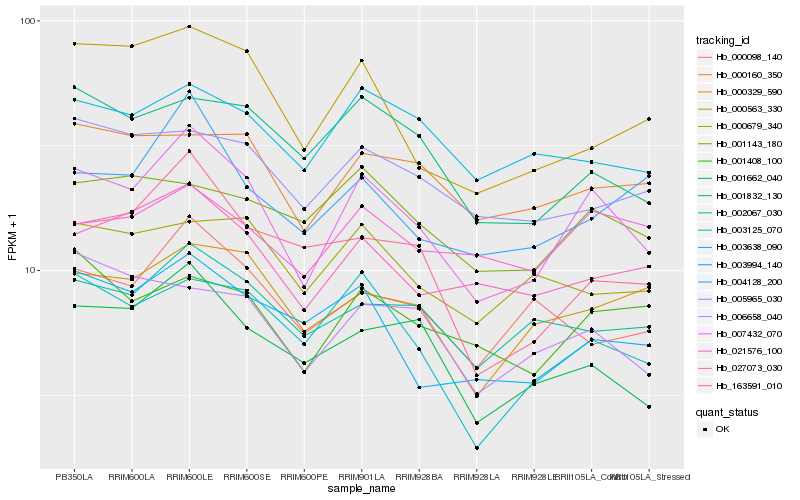
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007432_070 |
0.0 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 2 |
Hb_003125_070 |
0.110576912 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio1-like [Jatropha curcas] |
| 3 |
Hb_027073_030 |
0.1106064094 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 4 |
Hb_001662_040 |
0.111455217 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 5 |
Hb_002067_030 |
0.1128458408 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000563_330 |
0.1137147826 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_001832_130 |
0.1152840803 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 8 |
Hb_000329_590 |
0.1159484467 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_003994_140 |
0.116897361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001408_100 |
0.1170611161 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 11 |
Hb_000160_350 |
0.118705894 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 12 |
Hb_004128_200 |
0.1210090564 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 13 |
Hb_003638_090 |
0.1217042512 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 14 |
Hb_006658_040 |
0.1225461574 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 15 |
Hb_005965_030 |
0.1235438539 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 16 |
Hb_000098_140 |
0.1238686625 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 17 |
Hb_000679_340 |
0.1242480397 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 18 |
Hb_021576_100 |
0.1252009349 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 19 |
Hb_001143_180 |
0.1252422784 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 20 |
Hb_163591_010 |
0.1254591635 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |