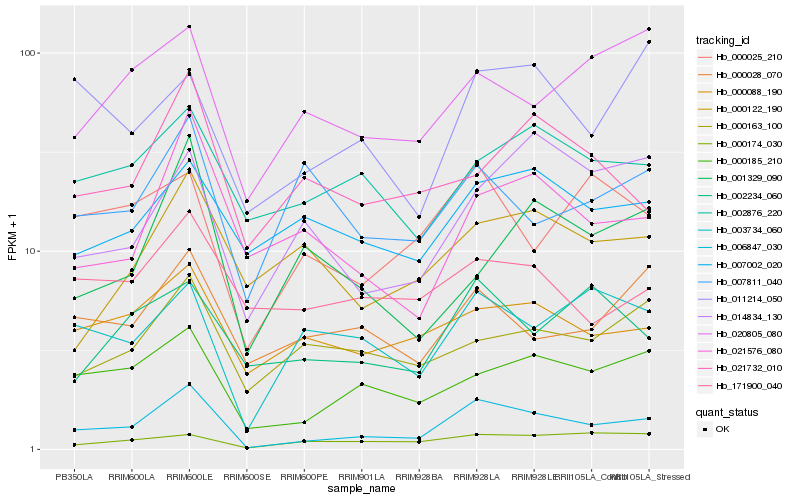
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006847_030 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000185_210 |
0.1746196319 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 3 |
Hb_021576_080 |
0.1944431445 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 4 |
Hb_000088_190 |
0.2015087538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003734_060 |
0.2038629539 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000028_070 |
0.2105457419 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 7 |
Hb_000025_210 |
0.2126799788 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_002876_220 |
0.2138363362 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 9 |
Hb_007002_020 |
0.2171280342 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 10 |
Hb_000163_100 |
0.2182279048 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 11 |
Hb_014834_130 |
0.2185129652 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000174_030 |
0.2203057275 |
- |
- |
hypothetical protein RCOM_1077370 [Ricinus communis] |
| 13 |
Hb_020805_080 |
0.2204946895 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 14 |
Hb_021732_010 |
0.2205391492 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 15 |
Hb_002234_060 |
0.2209184086 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 16 |
Hb_171900_040 |
0.2217319912 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 17 |
Hb_011214_050 |
0.2217507174 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 18 |
Hb_001329_090 |
0.2220673656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000122_190 |
0.2230872339 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007811_040 |
0.2231457767 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 isoform X1 [Gossypium raimondii] |