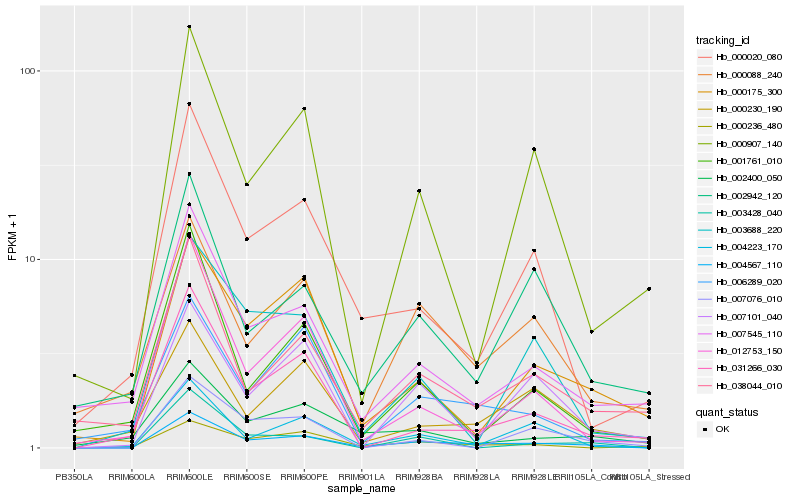
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004567_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s24610g [Populus trichocarpa] |
| 2 |
Hb_007545_110 |
0.1977477691 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_000020_080 |
0.1990059687 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 4 |
Hb_038044_010 |
0.1990511405 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 5 |
Hb_031266_030 |
0.2027342194 |
- |
- |
Kinase superfamily protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_004223_170 |
0.202760704 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 7 |
Hb_000175_300 |
0.2041941564 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 8 |
Hb_012753_150 |
0.2059074965 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 9 |
Hb_006289_020 |
0.2070336738 |
- |
- |
PREDICTED: shugoshin-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003428_040 |
0.2088571969 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000907_140 |
0.2144629903 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 12 |
Hb_000088_240 |
0.215282882 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000236_480 |
0.2164645415 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Jatropha curcas] |
| 14 |
Hb_007101_040 |
0.2181531844 |
- |
- |
PREDICTED: MLP-like protein 423 [Populus euphratica] |
| 15 |
Hb_002942_120 |
0.2210074355 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 16 |
Hb_001761_010 |
0.2210368473 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 17 |
Hb_003688_220 |
0.2228975556 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_002400_050 |
0.2229318247 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007076_010 |
0.224106952 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000230_190 |
0.2244945088 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |